The co-crystal C
10H
8N
2O
2·2C
7H
7NO
2 owes its formation to an intermolecular hydrogen bond between the O—H and N—O groups; the O

O distance is 2.5796 (17) Å. The dihedral angle between the planes of the pyridyl
N-oxide and aminobenzoic rings of the complex is 10.2 (1)°. An additional intermolecular C—H

O hydrogen bond is formed, giving rise to an eight-membered ring within the asymmetric unit. The structure is centrosymmetric about the mid-point of the bond joining the two pyridine rings. The crystal structure exhibits overlap between the aromatic rings of the molecules in the [
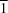
10] direction.
Supporting information
CCDC reference: 209962
Key indicators
- Single-crystal X-ray study
- T = 293 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.002 Å
(C-C) = 0.002 Å
- R factor = 0.044
- wR factor = 0.146
- Data-to-parameter ratio = 21.2
checkCIF results
No syntax errors found
ADDSYM reports no extra symmetry
Single yellow crystals of the DIPNO–PABA (1:2) [m.p. 453 (1) K] suitable for X-ray analysis were obtained by slow evaporation from an equimolecular solution of DIPNO and PABA in methanol. Initial reagents were purchased from Aldrich and were used without additional purification.
All H atoms, with the exception of the carboxyl atom HO2, were added at geometrically idealized positions and were allowed for as riding; C—H = 0.86–0.93 Å and Uiso(H)= 1.2Ueq of the carrier atom. Hydroxyl atom HO2 was located from a Fourier difference map and its coordinates were refined.
Data collection: CAD-4 EXPRESS (Enraf-Nonius, 1994); cell refinement: CAD-4 EXPRESS; data reduction: XCAD4 (Harms & Wocadlo, 1995); program(s) used to solve structure: SHELXS86 (Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEP-3 (Farrugia, 1997) and ZORTEP (Zsolnai, 1995); software used to prepare material for publication: SHELXL97.
Crystal data top
| C10H8N2O2·2C7H7NO2 | Dx = 1.383 Mg m−3 |
| Mr = 462.46 | Melting point: 453(1) K |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 25 reflections |
| a = 11.188 (1) Å | θ = 9–18° |
| b = 12.138 (2) Å | µ = 0.10 mm−1 |
| c = 16.354 (2) Å | T = 293 K |
| V = 2220.9 (5) Å3 | Regular block, yellow |
| Z = 4 | 0.22 × 0.18 × 0.12 mm |
| F(000) = 968 | |
Data collection top
Enraf-Nonius CAD-4
diffractometer | Rint = 0.026 |
| Radiation source: fine-focus sealed tube | θmax = 30.4°, θmin = 2.5° |
| Graphite monochromator | h = 0→15 |
| ω/2θ scans | k = 0→17 |
| 6757 measured reflections | l = −23→23 |
| 3350 independent reflections | 3 standard reflections every 120 min |
| 2057 reflections with I > 2σ(I) | intensity decay: 1.9% |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.146 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0639P)2 + 0.5535P]
where P = (Fo2 + 2Fc2)/3 |
| 3350 reflections | (Δ/σ)max < 0.001 |
| 158 parameters | Δρmax = 0.22 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Crystal data top
| C10H8N2O2·2C7H7NO2 | V = 2220.9 (5) Å3 |
| Mr = 462.46 | Z = 4 |
| Orthorhombic, Pbca | Mo Kα radiation |
| a = 11.188 (1) Å | µ = 0.10 mm−1 |
| b = 12.138 (2) Å | T = 293 K |
| c = 16.354 (2) Å | 0.22 × 0.18 × 0.12 mm |
Data collection top
Enraf-Nonius CAD-4
diffractometer | Rint = 0.026 |
| 6757 measured reflections | 3 standard reflections every 120 min |
| 3350 independent reflections | intensity decay: 1.9% |
| 2057 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.044 | 0 restraints |
| wR(F2) = 0.146 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.02 | Δρmax = 0.22 e Å−3 |
| 3350 reflections | Δρmin = −0.20 e Å−3 |
| 158 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.00584 (14) | −0.05247 (12) | 0.29335 (7) | 0.0715 (4) | |
| N1 | 0.00521 (12) | −0.03725 (11) | 0.21317 (7) | 0.0498 (3) | |
| C1 | −0.06048 (16) | −0.10416 (14) | 0.16603 (10) | 0.0537 (4) | |
| H1 | −0.1044 | −0.1606 | 0.1900 | 0.064* | |
| C2 | −0.06367 (15) | −0.09039 (13) | 0.08308 (10) | 0.0506 (4) | |
| H2 | −0.1100 | −0.1378 | 0.0516 | 0.061* | |
| C3 | 0.00058 (12) | −0.00731 (11) | 0.04476 (8) | 0.0378 (3) | |
| C4 | 0.06638 (16) | 0.06063 (14) | 0.09572 (9) | 0.0548 (4) | |
| H4 | 0.1102 | 0.1182 | 0.0732 | 0.066* | |
| C5 | 0.06824 (17) | 0.04477 (16) | 0.17887 (10) | 0.0604 (5) | |
| H5 | 0.1136 | 0.0913 | 0.2117 | 0.072* | |
| N2 | 0.37505 (15) | 0.33076 (15) | 0.67973 (9) | 0.0688 (5) | |
| HN21 | 0.3506 | 0.3116 | 0.7275 | 0.083* | |
| HN22 | 0.4262 | 0.3832 | 0.6746 | 0.083* | |
| O2 | 0.13107 (12) | 0.03202 (10) | 0.41001 (7) | 0.0593 (3) | |
| HO2 | 0.105 (2) | 0.007 (2) | 0.3615 (14) | 0.084 (7)* | |
| O3 | 0.22365 (13) | 0.15164 (12) | 0.32868 (6) | 0.0683 (4) | |
| C6 | 0.33268 (14) | 0.27774 (14) | 0.61185 (9) | 0.0482 (4) | |
| C7 | 0.37289 (14) | 0.30819 (14) | 0.53363 (9) | 0.0491 (4) | |
| H7 | 0.4283 | 0.3648 | 0.5280 | 0.059* | |
| C8 | 0.33072 (13) | 0.25479 (13) | 0.46541 (8) | 0.0455 (3) | |
| H8 | 0.3582 | 0.2758 | 0.4140 | 0.055* | |
| C9 | 0.24761 (14) | 0.16980 (12) | 0.47180 (8) | 0.0422 (3) | |
| C10 | 0.20820 (15) | 0.13959 (13) | 0.54979 (9) | 0.0482 (3) | |
| H10 | 0.1530 | 0.0828 | 0.5554 | 0.058* | |
| C11 | 0.25001 (15) | 0.19266 (14) | 0.61827 (9) | 0.0512 (4) | |
| H11 | 0.2227 | 0.1714 | 0.6696 | 0.061* | |
| C12 | 0.20161 (14) | 0.11742 (13) | 0.39705 (8) | 0.0461 (3) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0988 (10) | 0.0777 (9) | 0.0379 (6) | −0.0287 (8) | 0.0023 (6) | −0.0031 (6) |
| N1 | 0.0583 (8) | 0.0512 (7) | 0.0400 (6) | −0.0064 (6) | 0.0022 (6) | −0.0072 (5) |
| C1 | 0.0625 (10) | 0.0504 (8) | 0.0481 (8) | −0.0175 (7) | 0.0051 (7) | −0.0051 (7) |
| C2 | 0.0547 (9) | 0.0506 (8) | 0.0466 (7) | −0.0167 (7) | −0.0004 (6) | −0.0084 (6) |
| C3 | 0.0344 (6) | 0.0367 (6) | 0.0422 (7) | 0.0006 (5) | −0.0002 (5) | −0.0077 (5) |
| C4 | 0.0657 (10) | 0.0548 (9) | 0.0440 (8) | −0.0244 (8) | −0.0024 (7) | −0.0052 (7) |
| C5 | 0.0735 (11) | 0.0638 (10) | 0.0439 (8) | −0.0277 (9) | −0.0039 (8) | −0.0089 (7) |
| N2 | 0.0718 (10) | 0.0901 (12) | 0.0447 (7) | −0.0115 (9) | 0.0033 (7) | −0.0134 (8) |
| O2 | 0.0802 (8) | 0.0535 (7) | 0.0442 (6) | −0.0097 (6) | −0.0040 (6) | 0.0031 (5) |
| O3 | 0.0837 (9) | 0.0850 (9) | 0.0362 (5) | −0.0195 (7) | −0.0002 (5) | 0.0044 (6) |
| C6 | 0.0464 (8) | 0.0574 (9) | 0.0407 (7) | 0.0109 (7) | 0.0005 (6) | −0.0038 (6) |
| C7 | 0.0467 (8) | 0.0544 (8) | 0.0463 (8) | 0.0009 (7) | 0.0005 (6) | 0.0023 (7) |
| C8 | 0.0465 (7) | 0.0513 (8) | 0.0386 (6) | 0.0071 (7) | 0.0033 (6) | 0.0078 (6) |
| C9 | 0.0462 (7) | 0.0442 (7) | 0.0362 (6) | 0.0104 (6) | −0.0003 (6) | 0.0037 (5) |
| C10 | 0.0537 (8) | 0.0485 (8) | 0.0424 (7) | 0.0015 (7) | 0.0029 (6) | 0.0064 (6) |
| C11 | 0.0583 (9) | 0.0585 (9) | 0.0368 (6) | 0.0065 (7) | 0.0065 (7) | 0.0046 (6) |
| C12 | 0.0500 (8) | 0.0489 (8) | 0.0394 (7) | 0.0082 (7) | −0.0003 (6) | 0.0032 (6) |
Geometric parameters (Å, º) top
| O1—N1 | 1.3243 (16) | C4—C5 | 1.373 (2) |
| O1—O2 | 2.5796 (17) | C4—H4 | 0.9300 |
| O2—HO2 | 0.90 (2) | N2—C6 | 1.368 (2) |
| N2—O3i | 2.975 (2) | O2—C12 | 1.320 (2) |
| N2—HN21 | 0.8600 | O3—C12 | 1.2180 (17) |
| N2—O1ii | 3.095 (2) | C6—C11 | 1.390 (2) |
| N2—HN22 | 0.8600 | C6—C7 | 1.405 (2) |
| C5—O3 | 3.273 (2) | C7—C8 | 1.374 (2) |
| C5—H5 | 0.9300 | C7—H7 | 0.9300 |
| N1—C1 | 1.3394 (19) | C8—C9 | 1.393 (2) |
| N1—C5 | 1.343 (2) | C8—H8 | 0.9300 |
| C1—C2 | 1.367 (2) | C9—C10 | 1.398 (2) |
| C1—H1 | 0.9300 | C9—C12 | 1.471 (2) |
| C2—C3 | 1.388 (2) | C10—C11 | 1.374 (2) |
| C2—H2 | 0.9300 | C10—H10 | 0.9300 |
| C3—C4 | 1.384 (2) | C11—H11 | 0.9300 |
| C3—C3iii | 1.475 (3) | | |
| | | |
| O1—N1—C1 | 119.23 (13) | C12—O2—HO2 | 108.8 (15) |
| O1—N1—C5 | 120.95 (13) | N2—C6—C11 | 121.24 (14) |
| O1—HO2—O2 | 157 (2) | N2—C6—C7 | 120.26 (16) |
| N2—HN21—O3i | 145.6 | C11—C6—C7 | 118.49 (14) |
| N2—HN22—O1ii | 153.8 | C8—C7—C6 | 120.34 (15) |
| C5—H5—O3 | 158.4 | C8—C7—H7 | 119.8 |
| C1—N1—C5 | 119.82 (13) | C6—C7—H7 | 119.8 |
| N1—C1—C2 | 120.75 (15) | C7—C8—C9 | 121.19 (13) |
| N1—C1—H1 | 119.6 | C7—C8—H8 | 119.4 |
| C2—C1—H1 | 119.6 | C9—C8—H8 | 119.4 |
| C1—C2—C3 | 121.55 (14) | C8—C9—C10 | 118.23 (13) |
| C1—C2—H2 | 119.2 | C8—C9—C12 | 119.44 (12) |
| C3—C2—H2 | 119.2 | C10—C9—C12 | 122.30 (14) |
| C4—C3—C2 | 115.87 (13) | C11—C10—C9 | 120.87 (15) |
| C5—C4—C3 | 121.37 (15) | C11—C10—H10 | 119.6 |
| C5—C4—H4 | 119.3 | C9—C10—H10 | 119.6 |
| C3—C4—H4 | 119.3 | C10—C11—C6 | 120.88 (14) |
| N1—C5—C4 | 120.64 (14) | C10—C11—H11 | 119.6 |
| N1—C5—H5 | 119.7 | C6—C11—H11 | 119.6 |
| C4—C5—H5 | 119.7 | O3—C12—O2 | 122.43 (15) |
| C6—N2—HN21 | 120.0 | O3—C12—C9 | 123.00 (15) |
| C6—N2—HN22 | 120.0 | O2—C12—C9 | 114.53 (13) |
| HN21—N2—HN22 | 120.0 | | |
| | | |
| O1—N1—C1—C2 | 179.64 (17) | C7—C8—C9—C10 | −0.3 (2) |
| C5—N1—C1—C2 | 0.4 (3) | C7—C8—C9—C12 | 177.87 (14) |
| N1—C1—C2—C3 | 0.1 (3) | C8—C9—C10—C11 | 0.3 (2) |
| C1—C2—C3—C4 | −0.6 (2) | C12—C9—C10—C11 | −177.81 (14) |
| C2—C3—C4—C5 | 0.8 (3) | C9—C10—C11—C6 | 0.0 (2) |
| O1—N1—C5—C4 | −179.45 (18) | N2—C6—C11—C10 | −179.78 (16) |
| C1—N1—C5—C4 | −0.2 (3) | C7—C6—C11—C10 | −0.2 (2) |
| C3—C4—C5—N1 | −0.4 (3) | C8—C9—C12—O3 | −7.4 (2) |
| N2—C6—C7—C8 | 179.79 (15) | C10—C9—C12—O3 | 170.65 (16) |
| C11—C6—C7—C8 | 0.2 (2) | C10—C9—C12—O2 | −7.3 (2) |
| C6—C7—C8—C9 | 0.0 (2) | C8—C9—C12—O2 | 174.67 (14) |
| Symmetry codes: (i) x, −y+1/2, z+1/2; (ii) x+1/2, −y+1/2, −z+1; (iii) −x, −y, −z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—HO2···O1 | 0.90 (2) | 1.73 (2) | 2.580 (2) | 157 (2) |
| N2—HN21···O3i | 0.86 | 2.23 | 2.975 (2) | 146 |
| N2—HN22···O1ii | 0.86 | 2.30 | 3.095 (2) | 154 |
| C5—H5···O3 | 0.93 | 2.39 | 3.273 (2) | 158 |
| Symmetry codes: (i) x, −y+1/2, z+1/2; (ii) x+1/2, −y+1/2, −z+1. |
Experimental details
| Crystal data |
| Chemical formula | C10H8N2O2·2C7H7NO2 |
| Mr | 462.46 |
| Crystal system, space group | Orthorhombic, Pbca |
| Temperature (K) | 293 |
| a, b, c (Å) | 11.188 (1), 12.138 (2), 16.354 (2) |
| V (Å3) | 2220.9 (5) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.10 |
| Crystal size (mm) | 0.22 × 0.18 × 0.12 |
| |
| Data collection |
| Diffractometer | Enraf-Nonius CAD-4
diffractometer |
| Absorption correction | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 6757, 3350, 2057 |
| Rint | 0.026 |
| (sin θ/λ)max (Å−1) | 0.712 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.044, 0.146, 1.02 |
| No. of reflections | 3350 |
| No. of parameters | 158 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.22, −0.20 |
Selected geometric parameters (Å, º) top| O1—N1 | 1.3243 (16) | C3—C3i | 1.475 (3) |
| | | |
| O1—N1—C1 | 119.23 (13) | O3—C12—O2 | 122.43 (15) |
| O1—N1—C5 | 120.95 (13) | O3—C12—C9 | 123.00 (15) |
| N2—C6—C7 | 120.26 (16) | O2—C12—C9 | 114.53 (13) |
| | | |
| C8—C9—C12—O2 | 174.67 (14) | | |
| Symmetry code: (i) −x, −y, −z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—HO2···O1 | 0.90 (2) | 1.73 (2) | 2.580 (2) | 157 (2) |
| N2—HN21···O3ii | 0.86 | 2.23 | 2.975 (2) | 146 |
| N2—HN22···O1iii | 0.86 | 2.30 | 3.095 (2) | 154 |
| C5—H5···O3 | 0.93 | 2.39 | 3.273 (2) | 158 |
| Symmetry codes: (ii) x, −y+1/2, z+1/2; (iii) x+1/2, −y+1/2, −z+1. |
 O distance is 2.5796 (17) Å. The dihedral angle between the planes of the pyridyl N-oxide and aminobenzoic rings of the complex is 10.2 (1)°. An additional intermolecular C—H
O distance is 2.5796 (17) Å. The dihedral angle between the planes of the pyridyl N-oxide and aminobenzoic rings of the complex is 10.2 (1)°. An additional intermolecular C—H O hydrogen bond is formed, giving rise to an eight-membered ring within the asymmetric unit. The structure is centrosymmetric about the mid-point of the bond joining the two pyridine rings. The crystal structure exhibits overlap between the aromatic rings of the molecules in the [
O hydrogen bond is formed, giving rise to an eight-membered ring within the asymmetric unit. The structure is centrosymmetric about the mid-point of the bond joining the two pyridine rings. The crystal structure exhibits overlap between the aromatic rings of the molecules in the [![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.002 Å
(C-C) = 0.002 Å
 journal menu
journal menu










![[Figure 1]](http://journals.iucr.org/e/issues/2003/04/00/ac6032/ac6032fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/e/issues/2003/04/00/ac6032/ac6032fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/e/issues/2003/04/00/ac6032/ac6032fig3thm.gif)




The title compound, (I), was investigated as part of a study on D—H···A hydrogen bonding in system containing 4,4'-bipyridyl N,N-dioxide and hydrogen-bond donors (Thaimattam et al., 1998). Four molecular structures similar to (I) were found in the Cambridge Structural Database (Allen, 2002), namely 4,4'-bipyridine N,N'-dioxide urea clathrate, 4,4'-bipyridine N,N'-dioxide thiourea clathrate and 4,4'-bipyridine N,N'-dioxide dihydrate clathrate (Thaimattam et al., 1998), and 4-aminobenzoic acid (Lai & Marsh, 1967), and they were used as references to analyze the structural characteristics of complex (I).
The complex is held together by a strong intermolecular hydrogen bond (Emsley, 1984) between the O—H group of 4-aminobenzoic acid (PABA) molecule and the N—O group of 4,4'-bipyridyl N,N-dioxide (DIPNO) molecule. In the crystal, two acid components related by a centre of symmetry are linked to a centrosymmetric 4,4'-bipyridyl N,N-dioxide not only by the short O—H···O bonds but also by longer C5—H5.·O3 hydrogen bonds, giving rise to an eight-membered ring within the asymmetric unit. This results in an approximately linear chaining of the three molecules. The formation of these hydrogen bonds is probably related to the rotation of the carboxylic acid group around the C9—C12 bond [the dihedral angle between this group and the C6–C11 ring being 7.5 (1)°] and to the approximate planarity of the whole centrosymmetric unit. Selected hydrogen-bond O—H···O, N—H.·O and C—H.·O interactions are presented in Table 2. A perspective view of the DIPNO–PABA (1:2) adduct showing the atomic numbering scheme is given in Fig. 1. The dihedral angle formed by the planes which essentially contain the rings of DIPNO and PABA molecules is 10.2 (1)°. The values of bond lengths and other internal parameters of DIPNO are very close to values of structures reported in the literature (Thaimattam et al., 1998). The central C3—C3i bond length [1.475 (3) Å; symmetry code: (i) −x, −y, −z] is shorter than a normal C—C single-bond distance, showing that this C—C bond has probably a little π-bond character. The presence of a strong O—H.·O and other relatively weak C—H.·O intermolecular hydrogen bonds induces some structural changes in the donor complex. Indeed, the C9—C12, C12—O2 and C12—O3 bond lengths change from 1.471 (2), 1.320 (2) and 1.218 (2) Å in (I) to 1.455 (4), 1.295 (3) and 1.248 (5) Å in the PABA molecule. There are no unusual short intermolecular bonds in the crystal structure. The molecules of PABA and DIPNO(1/2 − x, −y, z − 1/2) are overlapped along the [110] direction at a distance of 3.454 (2) Å (Fig. 2). Additionally, hydrogen-bond interactions between atom N2 with atoms O3ii and O1iii of neighboring molecules [symmetry codes: (ii) ?, 1/2 − y, 1/2 + z; (iii) 1/2 + x, 1/2 − y, 1 − z] (please complete symmetry code) are observed (Fig. 3).