One of the two independent Mn atoms of the polymeric title compound, [Mn
3(C
16H
10O
4)
3(H
2O)
2]
n, lies on a special position of 3 site symmetry and the other on a special position of
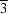
site symmetry. One end of the 4,4'-(ethene-1,2-diyl)dibenzoate dianion bridges the two independent Mn atoms. The water-coordinated Mn atom on the threefold axis is linked to the O atoms of three dianions in a tetrahedral environment. The Mn atom is linked to the O atoms of six different dianions in an octahedral geometry.
Supporting information
CCDC reference: 627788
Key indicators
- Single-crystal X-ray study
- T = 293 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (O-C) = 0.006 Å
(O-C) = 0.006 Å
- Disorder in main residue
- R factor = 0.069
- wR factor = 0.251
- Data-to-parameter ratio = 20.6
checkCIF/PLATON results
No syntax errors found
 Alert level A
PLAT242_ALERT_2_A Check Low Ueq as Compared to Neighbors for Mn1
Alert level A
PLAT242_ALERT_2_A Check Low Ueq as Compared to Neighbors for Mn1
| Author Response: ... The U(Mn) is normal but that of the coordinated water is large.
This water molecule is not hydrogen bonded to acceptor sites.
|
PLAT601_ALERT_2_A Structure Contains Solvent Accessible VOIDS of . 216.00 A 3
| Author Response: ... The structure is porous.
|
 Alert level B
PLAT301_ALERT_3_B Main Residue Disorder ......................... 39.00 Perc.
Alert level B
PLAT301_ALERT_3_B Main Residue Disorder ......................... 39.00 Perc.
 Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.661 0.892
Tmin(prime) and Tmax expected: 0.788 0.890
RR(prime) = 0.837
Please check that your absorption correction is appropriate.
RFACR01_ALERT_3_C The value of the weighted R factor is > 0.25
Weighted R factor given 0.251
PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ?
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 3.00 Ratio
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.83
PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ?
PLAT220_ALERT_2_C Large Non-Solvent O Ueq(max)/Ueq(min) ... 2.71 Ratio
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O2
PLAT710_ALERT_4_C Delete 1-2-3 or 2-3-4 Linear Torsion Angle ... # 4
O2 -MN2 -O2 -C1 12.00 0.00 10.555 1.555 1.555 1.555
Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.661 0.892
Tmin(prime) and Tmax expected: 0.788 0.890
RR(prime) = 0.837
Please check that your absorption correction is appropriate.
RFACR01_ALERT_3_C The value of the weighted R factor is > 0.25
Weighted R factor given 0.251
PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ?
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 3.00 Ratio
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.83
PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ?
PLAT220_ALERT_2_C Large Non-Solvent O Ueq(max)/Ueq(min) ... 2.71 Ratio
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O2
PLAT710_ALERT_4_C Delete 1-2-3 or 2-3-4 Linear Torsion Angle ... # 4
O2 -MN2 -O2 -C1 12.00 0.00 10.555 1.555 1.555 1.555
 Alert level G
FORMU01_ALERT_2_G There is a discrepancy between the atom counts in the
_chemical_formula_sum and the formula from the _atom_site* data.
Atom count from _chemical_formula_sum:C48 H34 Mn3 O14
Atom count from the _atom_site data: C48 H33.96 Mn3 O14
CELLZ01_ALERT_1_G Difference between formula and atom_site contents detected.
CELLZ01_ALERT_1_G ALERT: check formula stoichiometry or atom site occupancies.
From the CIF: _cell_formula_units_Z 3
From the CIF: _chemical_formula_sum C48 H34 Mn3 O14
TEST: Compare cell contents of formula and atom_site data
atom Z*formula cif sites diff
C 144.00 144.00 0.00
H 102.00 101.88 0.12
Mn 9.00 9.00 0.00
O 42.00 42.00 0.00
PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K
Alert level G
FORMU01_ALERT_2_G There is a discrepancy between the atom counts in the
_chemical_formula_sum and the formula from the _atom_site* data.
Atom count from _chemical_formula_sum:C48 H34 Mn3 O14
Atom count from the _atom_site data: C48 H33.96 Mn3 O14
CELLZ01_ALERT_1_G Difference between formula and atom_site contents detected.
CELLZ01_ALERT_1_G ALERT: check formula stoichiometry or atom site occupancies.
From the CIF: _cell_formula_units_Z 3
From the CIF: _chemical_formula_sum C48 H34 Mn3 O14
TEST: Compare cell contents of formula and atom_site data
atom Z*formula cif sites diff
C 144.00 144.00 0.00
H 102.00 101.88 0.12
Mn 9.00 9.00 0.00
O 42.00 42.00 0.00
PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K
2 ALERT level A = In general: serious problem
1 ALERT level B = Potentially serious problem
11 ALERT level C = Check and explain
4 ALERT level G = General alerts; check
7 ALERT type 1 CIF construction/syntax error, inconsistent or missing data
6 ALERT type 2 Indicator that the structure model may be wrong or deficient
4 ALERT type 3 Indicator that the structure quality may be low
1 ALERT type 4 Improvement, methodology, query or suggestion
0 ALERT type 5 Informative message, check
Data collection: RAPID-AUTO (Rigaku, 1998); cell refinement: RAPID-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2002); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: X-SEED (Barbour, 2001); software used to prepare material for publication: SHELXL97.
Poly[diaquatris[(
E)-4,4'-(ethene-1,2-diyl)dibenzoato]trimanganese(II)]
top
Crystal data top
| [Mn3(C16H10O4)3(H2O)2] | Dx = 1.280 Mg m−3 |
| Mr = 999.57 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, R3 | Cell parameters from 10113 reflections |
| Hall symbol: -R 3 | θ = 3.1–27.5° |
| a = 16.3366 (8) Å | µ = 0.78 mm−1 |
| c = 16.8286 (7) Å | T = 293 K |
| V = 3889.6 (2) Å3 | Block, yellow |
| Z = 3 | 0.30 × 0.22 × 0.15 mm |
| F(000) = 1527 | |
Data collection top
Rigaku R-AXIS RAPID IP
diffractometer | 1980 independent reflections |
| Radiation source: fine-focus sealed tube | 1663 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.021 |
| ω scans | θmax = 27.5°, θmin = 3.1° |
Absorption correction: multi-scan
(ABSCOR; Higashi, 1995) | h = −21→21 |
| Tmin = 0.661, Tmax = 0.892 | k = −21→21 |
| 12671 measured reflections | l = −20→21 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.069 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.251 | H-atom parameters constrained |
| S = 1.12 | w = 1/[σ2(Fo2) + (0.1598P)2 + 8.6886P]
where P = (Fo2 + 2Fc2)/3 |
| 1980 reflections | (Δ/σ)max = 0.001 |
| 96 parameters | Δρmax = 0.94 e Å−3 |
| 94 restraints | Δρmin = −0.58 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Mn1 | 0.0000 | 0.0000 | 0.20525 (6) | 0.0515 (4) | |
| Mn2 | 0.0000 | 0.0000 | 0.0000 | 0.0352 (4) | |
| O1 | −0.0113 (3) | 0.1201 (2) | 0.1910 (2) | 0.0728 (9) | |
| O2 | 0.0565 (2) | 0.1213 (2) | 0.0776 (2) | 0.0644 (8) | |
| O1w | 0.0000 | 0.0000 | 0.3285 (5) | 0.175 (6) | |
| H1W | −0.0027 | 0.0459 | 0.3448 | 0.209* | 0.33 |
| H2W | 0.0486 | 0.0027 | 0.3448 | 0.209* | 0.33 |
| C1 | 0.0197 (3) | 0.1546 (3) | 0.1232 (3) | 0.0562 (10) | |
| C2 | 0.0151 (10) | 0.2381 (7) | 0.0948 (8) | 0.0619 (12) | 0.50 |
| C3 | −0.0557 (10) | 0.2524 (8) | 0.1254 (7) | 0.067 (3) | 0.50 |
| H3 | −0.1010 | 0.2079 | 0.1593 | 0.081* | 0.50 |
| C4 | −0.0589 (8) | 0.3333 (8) | 0.1052 (7) | 0.093 (2) | 0.50 |
| H4 | −0.1063 | 0.3429 | 0.1256 | 0.112* | 0.50 |
| C5 | 0.0088 (7) | 0.3998 (6) | 0.0544 (6) | 0.089 (2) | 0.50 |
| C6 | 0.0796 (6) | 0.3854 (6) | 0.0238 (6) | 0.082 (3) | 0.50 |
| H6 | 0.1249 | 0.4299 | −0.0102 | 0.098* | 0.50 |
| C7 | 0.0828 (7) | 0.3046 (7) | 0.0440 (7) | 0.075 (3) | 0.50 |
| H7 | 0.1302 | 0.2950 | 0.0235 | 0.090* | 0.50 |
| C8 | 0.0007 (10) | 0.4825 (8) | 0.0363 (10) | 0.093 (4) | 0.50 |
| H8 | −0.0345 | 0.4954 | 0.0723 | 0.111* | 0.50 |
| C9 | −0.0115 (10) | 0.7608 (7) | −0.1007 (7) | 0.0619 (12) | 0.50 |
| C10 | 0.0439 (11) | 0.7353 (9) | −0.1452 (6) | 0.067 (3) | 0.50 |
| H10 | 0.0710 | 0.7665 | −0.1926 | 0.081* | 0.50 |
| C11 | 0.0588 (9) | 0.6631 (8) | −0.1190 (7) | 0.093 (2) | 0.50 |
| H11 | 0.0959 | 0.6460 | −0.1488 | 0.112* | 0.50 |
| C12 | 0.0183 (6) | 0.6164 (6) | −0.0482 (6) | 0.089 (2) | 0.50 |
| C13 | −0.0372 (6) | 0.6420 (6) | −0.0037 (5) | 0.082 (3) | 0.50 |
| H13 | −0.0643 | 0.6108 | 0.0436 | 0.098* | 0.50 |
| C14 | −0.0521 (8) | 0.7141 (7) | −0.0300 (7) | 0.075 (3) | 0.50 |
| H14 | −0.0891 | 0.7312 | −0.0002 | 0.090* | 0.50 |
| C15 | 0.0362 (10) | 0.5416 (8) | −0.0237 (11) | 0.093 (4) | 0.50 |
| H15 | 0.0792 | 0.5344 | −0.0550 | 0.111* | 0.50 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Mn1 | 0.0517 (5) | 0.0517 (5) | 0.0511 (6) | 0.0258 (2) | 0.000 | 0.000 |
| Mn2 | 0.0344 (5) | 0.0344 (5) | 0.0367 (6) | 0.0172 (2) | 0.000 | 0.000 |
| O1 | 0.104 (3) | 0.0564 (17) | 0.0649 (19) | 0.0447 (18) | −0.0082 (17) | −0.0063 (14) |
| O2 | 0.0582 (16) | 0.0481 (15) | 0.090 (2) | 0.0286 (13) | −0.0134 (15) | −0.0254 (14) |
| O1w | 0.233 (9) | 0.233 (9) | 0.058 (5) | 0.116 (5) | 0.000 | 0.000 |
| C1 | 0.061 (2) | 0.0435 (18) | 0.067 (2) | 0.0278 (17) | −0.0161 (18) | −0.0164 (17) |
| C2 | 0.081 (3) | 0.053 (2) | 0.062 (3) | 0.042 (2) | 0.003 (2) | −0.0041 (18) |
| C3 | 0.082 (4) | 0.062 (4) | 0.067 (6) | 0.043 (3) | 0.006 (4) | −0.008 (4) |
| C4 | 0.114 (4) | 0.088 (4) | 0.108 (5) | 0.073 (3) | 0.023 (4) | 0.006 (3) |
| C5 | 0.108 (6) | 0.085 (4) | 0.100 (4) | 0.067 (4) | 0.007 (4) | 0.008 (3) |
| C6 | 0.101 (6) | 0.071 (5) | 0.076 (5) | 0.044 (4) | 0.008 (4) | 0.011 (4) |
| C7 | 0.089 (6) | 0.076 (4) | 0.075 (4) | 0.053 (5) | 0.015 (4) | 0.003 (3) |
| C8 | 0.100 (9) | 0.085 (6) | 0.111 (6) | 0.059 (6) | 0.019 (6) | 0.010 (5) |
| C9 | 0.081 (3) | 0.053 (2) | 0.062 (3) | 0.042 (2) | 0.003 (2) | −0.0041 (18) |
| C10 | 0.082 (4) | 0.062 (4) | 0.067 (6) | 0.043 (3) | 0.006 (4) | −0.008 (4) |
| C11 | 0.114 (4) | 0.088 (4) | 0.108 (5) | 0.073 (3) | 0.023 (4) | 0.006 (3) |
| C12 | 0.108 (6) | 0.085 (4) | 0.100 (4) | 0.067 (4) | 0.007 (4) | 0.008 (3) |
| C13 | 0.101 (6) | 0.071 (5) | 0.076 (5) | 0.044 (4) | 0.008 (4) | 0.011 (4) |
| C14 | 0.089 (6) | 0.076 (4) | 0.075 (4) | 0.053 (5) | 0.015 (4) | 0.003 (3) |
| C15 | 0.100 (9) | 0.085 (6) | 0.111 (6) | 0.059 (6) | 0.019 (6) | 0.010 (5) |
Geometric parameters (Å, º) top
| Mn1—O1 | 2.075 (3) | C4—H4 | 0.9300 |
| Mn1—O1i | 2.075 (3) | C5—C6 | 1.3900 |
| Mn1—O1ii | 2.075 (3) | C5—C8 | 1.453 (6) |
| Mn1—O1w | 2.074 (9) | C6—C7 | 1.3900 |
| Mn2—O2 | 2.158 (3) | C6—H6 | 0.9300 |
| Mn2—O2i | 2.158 (3) | C7—H7 | 0.9300 |
| Mn2—O2ii | 2.158 (3) | C8—C15 | 1.315 (8) |
| Mn2—O2iii | 2.158 (3) | C8—H8 | 0.9300 |
| Mn2—O2iv | 2.158 (3) | C9—C10 | 1.3900 |
| Mn2—O2v | 2.158 (3) | C9—C14 | 1.3900 |
| O1—C1 | 1.261 (6) | C9—C1vi | 1.502 (8) |
| O2—C1 | 1.254 (5) | C10—C11 | 1.3900 |
| O1w—H1W | 0.8200 | C10—H10 | 0.9300 |
| O1w—H2W | 0.8200 | C11—C12 | 1.3900 |
| C1—C2 | 1.482 (9) | C11—H11 | 0.9300 |
| C1—C9vi | 1.502 (8) | C12—C13 | 1.3900 |
| C2—C3 | 1.3900 | C12—C15 | 1.453 (6) |
| C2—C7 | 1.3900 | C13—C14 | 1.3900 |
| C3—C4 | 1.3900 | C13—H13 | 0.9300 |
| C3—H3 | 0.9300 | C14—H14 | 0.9300 |
| C4—C5 | 1.3900 | C15—H15 | 0.9300 |
| | | |
| O1w—Mn1—O1i | 96.64 (10) | C3—C4—C5 | 120.0 |
| O1w—Mn1—O1 | 96.64 (10) | C3—C4—H4 | 120.0 |
| O1i—Mn1—O1 | 118.68 (4) | C5—C4—H4 | 120.0 |
| O1w—Mn1—O1ii | 96.64 (10) | C6—C5—C4 | 120.0 |
| O1i—Mn1—O1ii | 118.68 (4) | C6—C5—C8 | 123.3 (5) |
| O1—Mn1—O1ii | 118.68 (4) | C4—C5—C8 | 116.7 (5) |
| O2—Mn2—O2iii | 180.00 (17) | C5—C6—C7 | 120.0 |
| O2—Mn2—O2v | 92.85 (14) | C5—C6—H6 | 120.0 |
| O2iii—Mn2—O2v | 87.15 (14) | C7—C6—H6 | 120.0 |
| O2—Mn2—O2i | 87.15 (14) | C6—C7—C2 | 120.0 |
| O2iii—Mn2—O2i | 92.85 (14) | C6—C7—H7 | 120.0 |
| O2v—Mn2—O2i | 92.85 (14) | C2—C7—H7 | 120.0 |
| O2—Mn2—O2iv | 92.85 (14) | C15—C8—C5 | 129.1 (7) |
| O2iii—Mn2—O2iv | 87.15 (14) | C15—C8—H8 | 115.4 |
| O2v—Mn2—O2iv | 87.15 (14) | C5—C8—H8 | 115.4 |
| O2i—Mn2—O2iv | 180.0 | C10—C9—C14 | 120.0 |
| O2—Mn2—O2ii | 87.15 (14) | C10—C9—C1vi | 120.8 (9) |
| O2iii—Mn2—O2ii | 92.85 (14) | C14—C9—C1vi | 118.9 (9) |
| O2v—Mn2—O2ii | 180.00 (17) | C11—C10—C9 | 120.0 |
| O2i—Mn2—O2ii | 87.15 (14) | C11—C10—H10 | 120.0 |
| O2iv—Mn2—O2ii | 92.85 (14) | C9—C10—H10 | 120.0 |
| C1—O1—Mn1 | 108.7 (3) | C10—C11—C12 | 120.0 |
| C1—O2—Mn2 | 133.7 (3) | C10—C11—H11 | 120.0 |
| Mn1—O1w—H1W | 109.5 | C12—C11—H11 | 120.0 |
| Mn1—O1w—H2W | 109.5 | C13—C12—C11 | 120.0 |
| H1W—O1w—H2W | 109.5 | C13—C12—C15 | 122.4 (5) |
| O2—C1—O1 | 122.2 (4) | C11—C12—C15 | 117.6 (5) |
| O2—C1—C2 | 117.2 (6) | C14—C13—C12 | 120.0 |
| O1—C1—C2 | 120.6 (6) | C14—C13—H13 | 120.0 |
| O2—C1—C9vi | 121.8 (7) | C12—C13—H13 | 120.0 |
| O1—C1—C9vi | 116.0 (6) | C13—C14—C9 | 120.0 |
| C3—C2—C7 | 120.0 | C13—C14—H14 | 120.0 |
| C3—C2—C1 | 117.7 (9) | C9—C14—H14 | 120.0 |
| C7—C2—C1 | 122.1 (9) | C8—C15—C12 | 129.5 (8) |
| C2—C3—C4 | 120.0 | C8—C15—H15 | 115.3 |
| C2—C3—H3 | 120.0 | C12—C15—H15 | 115.3 |
| C4—C3—H3 | 120.0 | | |
| | | |
| O1w—Mn1—O1—C1 | 157.2 (3) | C3—C4—C5—C6 | 0.0 |
| O1i—Mn1—O1—C1 | −101.5 (3) | C3—C4—C5—C8 | −179.9 (3) |
| O1ii—Mn1—O1—C1 | 55.9 (4) | C4—C5—C6—C7 | 0.0 |
| O2iii—Mn2—O2—C1 | −123 (38) | C8—C5—C6—C7 | 179.8 (3) |
| O2v—Mn2—O2—C1 | 66.0 (4) | C5—C6—C7—C2 | 0.0 |
| O2i—Mn2—O2—C1 | −26.7 (5) | C3—C2—C7—C6 | 0.0 |
| O2iv—Mn2—O2—C1 | 153.3 (5) | C1—C2—C7—C6 | −175.0 (9) |
| O2ii—Mn2—O2—C1 | −114.0 (4) | C6—C5—C8—C15 | 20.6 (12) |
| Mn2—O2—C1—O1 | 78.4 (6) | C4—C5—C8—C15 | −159.6 (11) |
| Mn2—O2—C1—C2 | −102.4 (7) | C14—C9—C10—C11 | 0.0 |
| Mn2—O2—C1—C9vi | −102.6 (7) | C1vi—C9—C10—C11 | 173.2 (9) |
| Mn1—O1—C1—O2 | −4.5 (5) | C9—C10—C11—C12 | 0.0 |
| Mn1—O1—C1—C2 | 176.4 (6) | C10—C11—C12—C13 | 0.0 |
| Mn1—O1—C1—C9vi | 176.5 (6) | C10—C11—C12—C15 | −180.0 (3) |
| O2—C1—C2—C3 | 154.0 (5) | C11—C12—C13—C14 | 0.0 |
| O1—C1—C2—C3 | −26.9 (8) | C15—C12—C13—C14 | 180.0 (3) |
| C9vi—C1—C2—C3 | −28 (12) | C12—C13—C14—C9 | 0.0 |
| O2—C1—C2—C7 | −30.9 (9) | C10—C9—C14—C13 | 0.0 |
| O1—C1—C2—C7 | 148.3 (6) | C1vi—C9—C14—C13 | −173.3 (9) |
| C9vi—C1—C2—C7 | 147 (12) | C5—C8—C15—C12 | 170.6 (8) |
| C7—C2—C3—C4 | 0.0 | C13—C12—C15—C8 | 6.1 (12) |
| C1—C2—C3—C4 | 175.2 (9) | C11—C12—C15—C8 | −174.0 (11) |
| C2—C3—C4—C5 | 0.0 | | |
| Symmetry codes: (i) −y, x−y, z; (ii) −x+y, −x, z; (iii) −x, −y, −z; (iv) y, −x+y, −z; (v) x−y, x, −z; (vi) −x, −y+1, −z. |
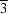 site symmetry. One end of the 4,4'-(ethene-1,2-diyl)dibenzoate dianion bridges the two independent Mn atoms. The water-coordinated Mn atom on the threefold axis is linked to the O atoms of three dianions in a tetrahedral environment. The Mn atom is linked to the O atoms of six different dianions in an octahedral geometry.
site symmetry. One end of the 4,4'-(ethene-1,2-diyl)dibenzoate dianion bridges the two independent Mn atoms. The water-coordinated Mn atom on the threefold axis is linked to the O atoms of three dianions in a tetrahedral environment. The Mn atom is linked to the O atoms of six different dianions in an octahedral geometry.![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (O-C) = 0.006 Å
(O-C) = 0.006 ÅAlert level A PLAT242_ALERT_2_A Check Low Ueq as Compared to Neighbors for Mn1
Alert level B PLAT301_ALERT_3_B Main Residue Disorder ......................... 39.00 Perc.
Alert level C ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90 Tmin and Tmax reported: 0.661 0.892 Tmin(prime) and Tmax expected: 0.788 0.890 RR(prime) = 0.837 Please check that your absorption correction is appropriate. RFACR01_ALERT_3_C The value of the weighted R factor is > 0.25 Weighted R factor given 0.251 PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ? PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ? PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 3.00 Ratio PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.83 PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ? PLAT220_ALERT_2_C Large Non-Solvent O Ueq(max)/Ueq(min) ... 2.71 Ratio PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1 PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O2 PLAT710_ALERT_4_C Delete 1-2-3 or 2-3-4 Linear Torsion Angle ... # 4 O2 -MN2 -O2 -C1 12.00 0.00 10.555 1.555 1.555 1.555
Alert level G FORMU01_ALERT_2_G There is a discrepancy between the atom counts in the _chemical_formula_sum and the formula from the _atom_site* data. Atom count from _chemical_formula_sum:C48 H34 Mn3 O14 Atom count from the _atom_site data: C48 H33.96 Mn3 O14 CELLZ01_ALERT_1_G Difference between formula and atom_site contents detected. CELLZ01_ALERT_1_G ALERT: check formula stoichiometry or atom site occupancies. From the CIF: _cell_formula_units_Z 3 From the CIF: _chemical_formula_sum C48 H34 Mn3 O14 TEST: Compare cell contents of formula and atom_site data atom Z*formula cif sites diff C 144.00 144.00 0.00 H 102.00 101.88 0.12 Mn 9.00 9.00 0.00 O 42.00 42.00 0.00 PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K

 journal menu
journal menu










 Alert level A
Alert level A
 Alert level B
Alert level B
 Alert level C
Alert level C
 Alert level G
Alert level G



