One of the two independent Cd atoms of the polymeric title compound, [Cd
3(C
16H
10O
4)
3(H
2O)
2]
n, lies on a special position of 3 site symmetry and the other on a special position of
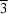
site symmetry. The compound adopts a carboxylate-bridged layer structure.
Supporting information
CCDC reference: 627789
Key indicators
- Single-crystal X-ray study
- T = 293 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (O-C) = 0.012 Å
(O-C) = 0.012 Å
- Disorder in main residue
- R factor = 0.064
- wR factor = 0.215
- Data-to-parameter ratio = 21.1
checkCIF/PLATON results
No syntax errors found
 Alert level A
PLAT242_ALERT_2_A Check Low Ueq as Compared to Neighbors for Cd1
Alert level A
PLAT242_ALERT_2_A Check Low Ueq as Compared to Neighbors for Cd1
| Author Response: ... The U(Mn) is normal but that of the coordinated water is large.
This water molecule is not hydrogen bonded to acceptor sites.
|
PLAT601_ALERT_2_A Structure Contains Solvent Accessible VOIDS of . 205.00 A 3
| Author Response: ... The structure is porous.
|
 Alert level B
PLAT301_ALERT_3_B Main Residue Disorder ......................... 39.00 Perc.
Alert level B
PLAT301_ALERT_3_B Main Residue Disorder ......................... 39.00 Perc.
 Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.601 0.807
Tmin(prime) and Tmax expected: 0.682 0.799
RR(prime) = 0.873
Please check that your absorption correction is appropriate.
PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ?
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 3.00 Ratio
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.86
PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ?
PLAT152_ALERT_1_C Supplied and Calc Volume s.u. Inconsistent ..... ?
PLAT220_ALERT_2_C Large Non-Solvent O Ueq(max)/Ueq(min) ... 2.73 Ratio
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O2
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for C4
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for C11
PLAT242_ALERT_2_C Check Low Ueq as Compared to Neighbors for C2
Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.601 0.807
Tmin(prime) and Tmax expected: 0.682 0.799
RR(prime) = 0.873
Please check that your absorption correction is appropriate.
PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ?
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 3.00 Ratio
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.86
PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ?
PLAT152_ALERT_1_C Supplied and Calc Volume s.u. Inconsistent ..... ?
PLAT220_ALERT_2_C Large Non-Solvent O Ueq(max)/Ueq(min) ... 2.73 Ratio
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O2
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for C4
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for C11
PLAT242_ALERT_2_C Check Low Ueq as Compared to Neighbors for C2
| Author Response: ... The U(Mn) is normal but that of the coordinated water is large.
This water molecule is not hydrogen bonded to acceptor sites.
|
PLAT242_ALERT_2_C Check Low Ueq as Compared to Neighbors for C9
| Author Response: ... The U(Mn) is normal but that of the coordinated water is large.
This water molecule is not hydrogen bonded to acceptor sites.
|
PLAT731_ALERT_1_C Bond Calc 2.205(11), Rep 2.204(5) ...... 2.20 su-Ra
CD1 -O1 1.555 3.555
 Alert level G
FORMU01_ALERT_2_G There is a discrepancy between the atom counts in the
_chemical_formula_sum and the formula from the _atom_site* data.
Atom count from _chemical_formula_sum:C48 H34 Cd3 O14
Atom count from the _atom_site data: C48 H33.96 Cd3 O14
CELLZ01_ALERT_1_G Difference between formula and atom_site contents detected.
CELLZ01_ALERT_1_G ALERT: check formula stoichiometry or atom site occupancies.
From the CIF: _cell_formula_units_Z 3
From the CIF: _chemical_formula_sum C48 H34 Cd3 O14
TEST: Compare cell contents of formula and atom_site data
atom Z*formula cif sites diff
C 144.00 144.00 0.00
H 102.00 101.88 0.12
Cd 9.00 9.00 0.00
O 42.00 42.00 0.00
PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K
Alert level G
FORMU01_ALERT_2_G There is a discrepancy between the atom counts in the
_chemical_formula_sum and the formula from the _atom_site* data.
Atom count from _chemical_formula_sum:C48 H34 Cd3 O14
Atom count from the _atom_site data: C48 H33.96 Cd3 O14
CELLZ01_ALERT_1_G Difference between formula and atom_site contents detected.
CELLZ01_ALERT_1_G ALERT: check formula stoichiometry or atom site occupancies.
From the CIF: _cell_formula_units_Z 3
From the CIF: _chemical_formula_sum C48 H34 Cd3 O14
TEST: Compare cell contents of formula and atom_site data
atom Z*formula cif sites diff
C 144.00 144.00 0.00
H 102.00 101.88 0.12
Cd 9.00 9.00 0.00
O 42.00 42.00 0.00
PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K
2 ALERT level A = In general: serious problem
1 ALERT level B = Potentially serious problem
15 ALERT level C = Check and explain
4 ALERT level G = General alerts; check
9 ALERT type 1 CIF construction/syntax error, inconsistent or missing data
10 ALERT type 2 Indicator that the structure model may be wrong or deficient
3 ALERT type 3 Indicator that the structure quality may be low
0 ALERT type 4 Improvement, methodology, query or suggestion
0 ALERT type 5 Informative message, check
Data collection: RAPID-AUTO (Rigaku, 1998); cell refinement: RAPID-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2002); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: X-SEED (Barbour, 2001); software used to prepare material for publication: SHELXL97.
Poly[diaquatris[(
E)-4,4'-(ethene-1,2-diyl)dibenzoato]tricadmium(II)]
top
Crystal data top
| [Cd3(C16H10O4)3(H2O)2] | Dx = 1.464 Mg m−3 |
| Mr = 1171.95 | Mo Kα radiation, λ = 0.71073 Å |
| Trigonal, R3 | Cell parameters from 7979 reflections |
| Hall symbol: -R 3 | θ = 3.1–27.4° |
| a = 16.4773 (7) Å | µ = 1.25 mm−1 |
| c = 16.9571 (8) Å | T = 293 K |
| V = 3987.1 (3) Å3 | Prism, colourless |
| Z = 3 | 0.30 × 0.24 × 0.18 mm |
| F(000) = 1734 | |
Data collection top
Rigaku R-AXIS RAPID IP
diffractometer | 2026 independent reflections |
| Radiation source: fine-focus sealed tube | 1335 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.054 |
| ω scans | θmax = 27.4°, θmin = 3.1° |
Absorption correction: multi-scan
(ABSCOR; Higashi, 1995) | h = −21→21 |
| Tmin = 0.601, Tmax = 0.807 | k = −19→21 |
| 13023 measured reflections | l = −21→21 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.064 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.215 | H-atom parameters constrained |
| S = 1.16 | w = 1/[σ2(Fo2) + (0.1146P)2 + 13.9612P]
where P = (Fo2 + 2Fc2)/3 |
| 2026 reflections | (Δ/σ)max = 0.001 |
| 96 parameters | Δρmax = 0.72 e Å−3 |
| 94 restraints | Δρmin = −1.32 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cd1 | 0.0000 | 0.0000 | 0.20467 (6) | 0.0560 (4) | |
| Cd2 | 0.0000 | 0.0000 | 0.0000 | 0.0453 (4) | |
| O1 | −0.0182 (5) | 0.1231 (4) | 0.1915 (4) | 0.0778 (17) | |
| O2 | 0.0552 (4) | 0.1247 (4) | 0.0834 (4) | 0.0697 (15) | |
| O1W | 0.0000 | 0.0000 | 0.3358 (9) | 0.192 (9) | |
| H1W | 0.0264 | 0.0542 | 0.3519 | 0.230* | 0.33 |
| H2W | 0.0277 | −0.0264 | 0.3519 | 0.230* | 0.33 |
| C1 | 0.0154 (6) | 0.1568 (5) | 0.1255 (5) | 0.0616 (18) | |
| C2 | 0.0243 (9) | 0.2478 (8) | 0.1048 (9) | 0.055 (3) | 0.50 |
| C3 | −0.0399 (10) | 0.2676 (10) | 0.1401 (9) | 0.079 (4) | 0.50 |
| H3 | −0.0797 | 0.2285 | 0.1794 | 0.095* | 0.50 |
| C4 | −0.0449 (9) | 0.3459 (10) | 0.1167 (9) | 0.100 (4) | 0.50 |
| H4 | −0.0879 | 0.3592 | 0.1403 | 0.120* | 0.50 |
| C5 | 0.0145 (9) | 0.4043 (7) | 0.0580 (8) | 0.089 (4) | 0.50 |
| C6 | 0.0788 (9) | 0.3845 (9) | 0.0227 (8) | 0.095 (5) | 0.50 |
| H6 | 0.1185 | 0.4236 | −0.0166 | 0.114* | 0.50 |
| C7 | 0.0837 (9) | 0.3062 (9) | 0.0461 (9) | 0.076 (4) | 0.50 |
| H7 | 0.1267 | 0.2929 | 0.0225 | 0.091* | 0.50 |
| C8 | 0.0059 (14) | 0.4838 (10) | 0.0364 (13) | 0.111 (6) | 0.50 |
| H8 | −0.0320 | 0.4945 | 0.0704 | 0.134* | 0.50 |
| C9 | 0.0027 (9) | 0.7681 (8) | −0.0937 (9) | 0.055 (3) | 0.50 |
| C10 | 0.0648 (11) | 0.7484 (10) | −0.1333 (9) | 0.079 (4) | 0.50 |
| H10 | 0.0985 | 0.7847 | −0.1762 | 0.095* | 0.50 |
| C11 | 0.0764 (10) | 0.6744 (10) | −0.1086 (9) | 0.100 (4) | 0.50 |
| H11 | 0.1179 | 0.6612 | −0.1350 | 0.120* | 0.50 |
| C12 | 0.0260 (8) | 0.6201 (7) | −0.0444 (8) | 0.089 (4) | 0.50 |
| C13 | −0.0360 (9) | 0.6399 (9) | −0.0048 (8) | 0.095 (5) | 0.50 |
| H13 | −0.0698 | 0.6036 | 0.0381 | 0.114* | 0.50 |
| C14 | −0.0477 (9) | 0.7139 (9) | −0.0295 (9) | 0.076 (4) | 0.50 |
| H14 | −0.0892 | 0.7271 | −0.0031 | 0.091* | 0.50 |
| C15 | 0.0407 (13) | 0.5446 (10) | −0.0216 (13) | 0.111 (6) | 0.50 |
| H15 | 0.0825 | 0.5375 | −0.0537 | 0.134* | 0.50 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cd1 | 0.0552 (4) | 0.0552 (4) | 0.0575 (6) | 0.0276 (2) | 0.000 | 0.000 |
| Cd2 | 0.0453 (4) | 0.0453 (4) | 0.0453 (6) | 0.0226 (2) | 0.000 | 0.000 |
| O1 | 0.116 (5) | 0.062 (3) | 0.070 (4) | 0.055 (3) | 0.002 (3) | 0.004 (3) |
| O2 | 0.072 (3) | 0.052 (3) | 0.095 (4) | 0.038 (3) | −0.013 (3) | −0.025 (3) |
| O1W | 0.251 (15) | 0.251 (15) | 0.073 (10) | 0.126 (8) | 0.000 | 0.000 |
| C1 | 0.072 (5) | 0.049 (4) | 0.068 (5) | 0.034 (4) | −0.012 (4) | −0.010 (4) |
| C2 | 0.054 (7) | 0.053 (5) | 0.053 (5) | 0.022 (5) | −0.013 (5) | −0.011 (4) |
| C3 | 0.083 (7) | 0.074 (6) | 0.094 (6) | 0.048 (5) | 0.007 (5) | 0.000 (4) |
| C4 | 0.100 (8) | 0.096 (7) | 0.117 (7) | 0.059 (6) | 0.008 (6) | 0.002 (5) |
| C5 | 0.098 (8) | 0.083 (6) | 0.104 (7) | 0.058 (6) | −0.008 (6) | 0.000 (5) |
| C6 | 0.106 (8) | 0.088 (7) | 0.091 (7) | 0.047 (6) | 0.004 (6) | 0.009 (5) |
| C7 | 0.084 (7) | 0.074 (6) | 0.079 (6) | 0.047 (6) | 0.007 (5) | −0.001 (5) |
| C8 | 0.124 (11) | 0.099 (9) | 0.123 (10) | 0.064 (8) | −0.005 (8) | 0.003 (7) |
| C9 | 0.054 (7) | 0.053 (5) | 0.053 (5) | 0.022 (5) | −0.013 (5) | −0.011 (4) |
| C10 | 0.083 (7) | 0.074 (6) | 0.094 (6) | 0.048 (5) | 0.007 (5) | 0.000 (4) |
| C11 | 0.100 (8) | 0.096 (7) | 0.117 (7) | 0.059 (6) | 0.008 (6) | 0.002 (5) |
| C12 | 0.098 (8) | 0.083 (6) | 0.104 (7) | 0.058 (6) | −0.008 (6) | 0.000 (5) |
| C13 | 0.106 (8) | 0.088 (7) | 0.091 (7) | 0.047 (6) | 0.004 (6) | 0.009 (5) |
| C14 | 0.084 (7) | 0.074 (6) | 0.079 (6) | 0.047 (6) | 0.007 (5) | −0.001 (5) |
| C15 | 0.124 (11) | 0.099 (9) | 0.123 (10) | 0.064 (8) | −0.005 (8) | 0.003 (7) |
Geometric parameters (Å, º) top
| Cd1—O1 | 2.204 (5) | C4—H4 | 0.9300 |
| Cd1—O1i | 2.204 (5) | C5—C6 | 1.3900 |
| Cd1—O1ii | 2.204 (5) | C5—C8 | 1.433 (6) |
| Cd1—O1w | 2.22 (2) | C6—C7 | 1.3900 |
| Cd2—O2 | 2.277 (5) | C6—H6 | 0.9300 |
| Cd2—O2i | 2.277 (5) | C7—H7 | 0.9300 |
| Cd2—O2ii | 2.277 (5) | C8—C15 | 1.314 (9) |
| Cd2—O2iii | 2.277 (5) | C8—H8 | 0.9300 |
| Cd2—O2iv | 2.277 (5) | C9—C10 | 1.3900 |
| Cd2—O2v | 2.277 (5) | C9—C14 | 1.3900 |
| O1—C1 | 1.250 (9) | C9—C1vi | 1.509 (8) |
| O2—C1 | 1.251 (9) | C10—C11 | 1.3900 |
| O1W—H1W | 0.8200 | C10—H10 | 0.9300 |
| O1W—H2W | 0.8200 | C11—C12 | 1.3900 |
| C1—C2 | 1.473 (13) | C11—H11 | 0.9300 |
| C1—C9vi | 1.509 (8) | C12—C13 | 1.3900 |
| C2—C3 | 1.3900 | C12—C15 | 1.435 (6) |
| C2—C7 | 1.3900 | C13—C14 | 1.3900 |
| C3—C4 | 1.3900 | C13—H13 | 0.9300 |
| C3—H3 | 0.9300 | C14—H14 | 0.9300 |
| C4—C5 | 1.3900 | C15—H15 | 0.9300 |
| | | |
| O1ii—Cd1—O1i | 118.99 (6) | C3—C4—C5 | 120.0 |
| O1ii—Cd1—O1 | 118.99 (6) | C3—C4—H4 | 120.0 |
| O1i—Cd1—O1 | 118.99 (6) | C5—C4—H4 | 120.0 |
| O1ii—Cd1—O1W | 95.81 (16) | C6—C5—C4 | 120.0 |
| O1i—Cd1—O1W | 95.81 (16) | C6—C5—C8 | 122.7 (5) |
| O1—Cd1—O1W | 95.81 (16) | C4—C5—C8 | 117.3 (5) |
| O2—Cd2—O2ii | 85.5 (2) | C5—C6—C7 | 120.0 |
| O2—Cd2—O2iii | 180.0 (3) | C5—C6—H6 | 120.0 |
| O2ii—Cd2—O2iii | 94.5 (2) | C7—C6—H6 | 120.0 |
| O2—Cd2—O2iv | 94.5 (2) | C6—C7—C2 | 120.0 |
| O2ii—Cd2—O2iv | 94.5 (2) | C6—C7—H7 | 120.0 |
| O2iii—Cd2—O2iv | 85.5 (2) | C2—C7—H7 | 120.0 |
| O2—Cd2—O2i | 85.5 (2) | C15—C8—C5 | 133.1 (10) |
| O2ii—Cd2—O2i | 85.5 (2) | C15—C8—H8 | 113.4 |
| O2iii—Cd2—O2i | 94.5 (2) | C5—C8—H8 | 113.4 |
| O2iv—Cd2—O2i | 180.0 (3) | C10—C9—C14 | 120.0 |
| O2—Cd2—O2v | 94.5 (2) | C10—C9—C1vi | 119.3 (10) |
| O2ii—Cd2—O2v | 180.0 (3) | C14—C9—C1vi | 120.5 (10) |
| O2iii—Cd2—O2v | 85.5 (2) | C9—C10—C11 | 120.0 |
| O2iv—Cd2—O2v | 85.5 (2) | C9—C10—H10 | 120.0 |
| O2i—Cd2—O2v | 94.5 (2) | C11—C10—H10 | 120.0 |
| C1—O1—Cd1 | 105.5 (5) | C12—C11—C10 | 120.0 |
| C1—O2—Cd2 | 132.8 (5) | C12—C11—H11 | 120.0 |
| Cd1—O1W—H1W | 109.5 | C10—C11—H11 | 120.0 |
| Cd1—O1W—H2W | 109.5 | C13—C12—C11 | 120.0 |
| H1W—O1W—H2W | 109.5 | C13—C12—C15 | 122.4 (5) |
| O1—C1—O2 | 122.1 (7) | C11—C12—C15 | 117.6 (5) |
| O1—C1—C2 | 117.6 (9) | C12—C13—C14 | 120.0 |
| O2—C1—C2 | 119.2 (9) | C12—C13—H13 | 120.0 |
| O1—C1—C9vi | 117.3 (9) | C14—C13—H13 | 120.0 |
| O2—C1—C9vi | 120.2 (9) | C13—C14—C9 | 120.0 |
| C3—C2—C7 | 120.0 | C13—C14—H14 | 120.0 |
| C3—C2—C1 | 115.9 (10) | C9—C14—H14 | 120.0 |
| C7—C2—C1 | 123.5 (10) | C8—C15—C12 | 131.7 (10) |
| C2—C3—C4 | 120.0 | C8—C15—H15 | 114.1 |
| C2—C3—H3 | 120.0 | C12—C15—H15 | 114.1 |
| C4—C3—H3 | 120.0 | | |
| | | |
| O1ii—Cd1—O1—C1 | 55.0 (6) | C3—C4—C5—C6 | 0.0 |
| O1i—Cd1—O1—C1 | −105.1 (5) | C3—C4—C5—C8 | 179.9 (3) |
| O1W—Cd1—O1—C1 | 155.0 (5) | C4—C5—C6—C7 | 0.0 |
| O2ii—Cd2—O2—C1 | −117.9 (6) | C8—C5—C6—C7 | −179.9 (3) |
| O2iv—Cd2—O2—C1 | 147.8 (8) | C5—C6—C7—C2 | 0.0 |
| O2i—Cd2—O2—C1 | −32.2 (8) | C3—C2—C7—C6 | 0.0 |
| O2v—Cd2—O2—C1 | 62.1 (6) | C1—C2—C7—C6 | 171.1 (11) |
| Cd1—O1—C1—O2 | −4.3 (9) | C6—C5—C8—C15 | 10.2 (19) |
| Cd1—O1—C1—C2 | −172.1 (7) | C4—C5—C8—C15 | −169.8 (19) |
| Cd1—O1—C1—C9vi | 169.2 (7) | C14—C9—C10—C11 | 0.0 |
| Cd2—O2—C1—O1 | 81.3 (10) | C1vi—C9—C10—C11 | −174.7 (10) |
| Cd2—O2—C1—C2 | −111.1 (9) | C9—C10—C11—C12 | 0.0 |
| Cd2—O2—C1—C9vi | −92.1 (10) | C10—C11—C12—C13 | 0.0 |
| O1—C1—C2—C3 | −28.2 (11) | C10—C11—C12—C15 | 180.0 (3) |
| O2—C1—C2—C3 | 163.7 (8) | C11—C12—C13—C14 | 0.0 |
| C9vi—C1—C2—C3 | 65 (4) | C15—C12—C13—C14 | −180.0 (3) |
| O1—C1—C2—C7 | 160.3 (9) | C12—C13—C14—C9 | 0.0 |
| O2—C1—C2—C7 | −7.8 (14) | C10—C9—C14—C13 | 0.0 |
| C9vi—C1—C2—C7 | −106 (4) | C1vi—C9—C14—C13 | 174.6 (10) |
| C7—C2—C3—C4 | 0.0 | C5—C8—C15—C12 | 175.4 (9) |
| C1—C2—C3—C4 | −171.8 (10) | C13—C12—C15—C8 | −1.9 (18) |
| C2—C3—C4—C5 | 0.0 | C11—C12—C15—C8 | 178.2 (18) |
| Symmetry codes: (i) −y, x−y, z; (ii) −x+y, −x, z; (iii) −x, −y, −z; (iv) y, −x+y, −z; (v) x−y, x, −z; (vi) −x, −y+1, −z. |
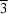 site symmetry. The compound adopts a carboxylate-bridged layer structure.
site symmetry. The compound adopts a carboxylate-bridged layer structure.![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (O-C) = 0.012 Å
(O-C) = 0.012 ÅAlert level A PLAT242_ALERT_2_A Check Low Ueq as Compared to Neighbors for Cd1
Alert level B PLAT301_ALERT_3_B Main Residue Disorder ......................... 39.00 Perc.
Alert level C ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90 Tmin and Tmax reported: 0.601 0.807 Tmin(prime) and Tmax expected: 0.682 0.799 RR(prime) = 0.873 Please check that your absorption correction is appropriate. PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ? PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ? PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 3.00 Ratio PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.86 PLAT068_ALERT_1_C Reported F000 Differs from Calcd (or Missing)... ? PLAT152_ALERT_1_C Supplied and Calc Volume s.u. Inconsistent ..... ? PLAT220_ALERT_2_C Large Non-Solvent O Ueq(max)/Ueq(min) ... 2.73 Ratio PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1 PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O2 PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for C4 PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for C11 PLAT242_ALERT_2_C Check Low Ueq as Compared to Neighbors for C2
Alert level G FORMU01_ALERT_2_G There is a discrepancy between the atom counts in the _chemical_formula_sum and the formula from the _atom_site* data. Atom count from _chemical_formula_sum:C48 H34 Cd3 O14 Atom count from the _atom_site data: C48 H33.96 Cd3 O14 CELLZ01_ALERT_1_G Difference between formula and atom_site contents detected. CELLZ01_ALERT_1_G ALERT: check formula stoichiometry or atom site occupancies. From the CIF: _cell_formula_units_Z 3 From the CIF: _chemical_formula_sum C48 H34 Cd3 O14 TEST: Compare cell contents of formula and atom_site data atom Z*formula cif sites diff C 144.00 144.00 0.00 H 102.00 101.88 0.12 Cd 9.00 9.00 0.00 O 42.00 42.00 0.00 PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K

 journal menu
journal menu










 Alert level A
Alert level A
 Alert level B
Alert level B
 Alert level C
Alert level C
 Alert level G
Alert level G



