The metal atom in the title compound, [Cu(C
7H
5O
3)
2(C
3H
4N
2)
2], is coordinated by two O atoms (from two carboxylate anions) and two N atoms (from two
N-heterocycles) in a square-planar geometry; it occupies a special position of site symmetry
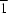
. Above and below the square are the hydroxy O atoms of adjacent molecules. This weak interaction [2.646 (2) Å] leads to the formation of a linear chain; the chains are consolidated into a layer through hydrogen bonds.
Supporting information
CCDC reference: 630532
Key indicators
- Single-crystal X-ray study
- T = 295 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.004 Å
(C-C) = 0.004 Å
- R factor = 0.041
- wR factor = 0.132
- Data-to-parameter ratio = 15.2
checkCIF/PLATON results
No syntax errors found
 Alert level C
PLAT094_ALERT_2_C Ratio of Maximum / Minimum Residual Density .... 2.22
PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
O3 -H3O 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
N2 -H2N 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
O3 -H3O 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
N2 -H2N 1.555 1.555
PLAT736_ALERT_1_C H...A Calc 1.76(3), Rep 1.760(10) ...... 3.00 su-Ra
H3O -O2 1.555 1.556
Alert level C
PLAT094_ALERT_2_C Ratio of Maximum / Minimum Residual Density .... 2.22
PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
O3 -H3O 1.555 1.555
PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
N2 -H2N 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
O3 -H3O 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra
N2 -H2N 1.555 1.555
PLAT736_ALERT_1_C H...A Calc 1.76(3), Rep 1.760(10) ...... 3.00 su-Ra
H3O -O2 1.555 1.556
0 ALERT level A = In general: serious problem
0 ALERT level B = Potentially serious problem
6 ALERT level C = Check and explain
0 ALERT level G = General alerts; check
5 ALERT type 1 CIF construction/syntax error, inconsistent or missing data
1 ALERT type 2 Indicator that the structure model may be wrong or deficient
0 ALERT type 3 Indicator that the structure quality may be low
0 ALERT type 4 Improvement, methodology, query or suggestion
0 ALERT type 5 Informative message, check
Data collection: APEX2 software (Bruker, 2004); cell refinement: SAINT (Bruker, 2004); data reduction: SAINT; program(s) used to solve structure: Difference Fourier synthesis, with Cu1 at (1/2, 1/2, 1/2); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: X-SEED (Barbour, 2001); software used to prepare material for publication: publCIF (Westrip, 2006).
Crystal data top
| [Cu(C7H5O3)2(C3H4N2)2] | F(000) = 486 |
| Mr = 473.92 | Dx = 1.573 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3090 reflections |
| a = 9.299 (1) Å | θ = 2.7–29.0° |
| b = 13.333 (1) Å | µ = 1.14 mm−1 |
| c = 8.078 (1) Å | T = 295 K |
| β = 92.196 (1)° | Block, blue |
| V = 1000.80 (18) Å3 | 0.32 × 0.22 × 0.13 mm |
| Z = 2 | |
Data collection top
Bruker APEX2 area-detector
diffractometer | 2281 independent reflections |
| Radiation source: fine-focus sealed tube | 1859 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.047 |
| φ and ω scans | θmax = 27.5°, θmin = 2.7° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 1996) | h = −12→11 |
| Tmin = 0.712, Tmax = 0.866 | k = −10→17 |
| 6475 measured reflections | l = −10→10 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.132 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.09 | w = 1/[σ2(Fo2) + (0.0767P)2 + 0.451P]
where P = (Fo2 + 2Fc2)/3 |
| 2281 reflections | (Δ/σ)max = 0.001 |
| 150 parameters | Δρmax = 1.00 e Å−3 |
| 2 restraints | Δρmin = −0.45 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cu1 | 0.5000 | 0.5000 | 0.5000 | 0.0278 (2) | |
| O1 | 0.6093 (2) | 0.5471 (1) | 0.7003 (2) | 0.0332 (4) | |
| O2 | 0.8294 (2) | 0.5635 (2) | 0.6012 (2) | 0.0518 (6) | |
| O3 | 0.7265 (2) | 0.5086 (2) | 1.3120 (3) | 0.0397 (5) | |
| N1 | 0.5549 (2) | 0.3608 (2) | 0.5588 (3) | 0.0310 (5) | |
| N2 | 0.5375 (3) | 0.2049 (2) | 0.6409 (3) | 0.0407 (6) | |
| C1 | 0.7452 (3) | 0.5632 (2) | 0.7165 (3) | 0.0303 (5) | |
| C2 | 0.8028 (3) | 0.5819 (2) | 0.8897 (3) | 0.0266 (5) | |
| C3 | 0.7332 (3) | 0.5418 (2) | 1.0247 (3) | 0.0277 (5) | |
| C4 | 0.7913 (3) | 0.5551 (2) | 1.1839 (3) | 0.0281 (5) | |
| C5 | 0.9152 (3) | 0.6114 (2) | 1.2093 (3) | 0.0392 (7) | |
| C6 | 0.9825 (4) | 0.6523 (3) | 1.0748 (4) | 0.0486 (8) | |
| C7 | 0.9288 (3) | 0.6365 (2) | 0.9157 (3) | 0.0394 (7) | |
| C8 | 0.6895 (3) | 0.3247 (2) | 0.6034 (4) | 0.0436 (7) | |
| C9 | 0.6793 (3) | 0.2285 (2) | 0.6554 (4) | 0.0449 (7) | |
| C10 | 0.4670 (3) | 0.2858 (2) | 0.5827 (4) | 0.0396 (6) | |
| H3 | 0.6479 | 0.5061 | 1.0079 | 0.033* | |
| H5 | 0.9531 | 0.6218 | 1.3161 | 0.047* | |
| H6 | 1.0651 | 0.6908 | 1.0922 | 0.058* | |
| H7 | 0.9767 | 0.6622 | 0.8262 | 0.047* | |
| H8 | 0.7747 | 0.3610 | 0.5986 | 0.052* | |
| H9 | 0.7541 | 0.1872 | 0.6933 | 0.054* | |
| H10 | 0.3681 | 0.2885 | 0.5619 | 0.047* | |
| H3o | 0.770 (4) | 0.525 (3) | 1.402 (3) | 0.06 (1)* | |
| H2n | 0.495 (4) | 0.152 (2) | 0.673 (5) | 0.06 (1)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.0357 (3) | 0.0247 (3) | 0.0220 (3) | 0.0007 (2) | −0.0136 (2) | −0.0012 (2) |
| O1 | 0.038 (1) | 0.036 (1) | 0.024 (1) | 0.003 (1) | −0.014 (1) | −0.006 (1) |
| O2 | 0.056 (1) | 0.083 (2) | 0.017 (1) | −0.014 (1) | 0.000 (1) | −0.004 (1) |
| O3 | 0.043 (1) | 0.056 (1) | 0.020 (1) | −0.011 (1) | 0.00 (1) | 0.004 (1) |
| N1 | 0.037 (1) | 0.028 (1) | 0.027 (1) | 0.001 (1) | −0.006 (1) | 0.002 (1) |
| N2 | 0.046 (1) | 0.031 (1) | 0.045 (1) | −0.002 (1) | 0.002 (1) | 0.009 (1) |
| C1 | 0.041 (1) | 0.031 (1) | 0.019 (1) | −0.002 (1) | −0.007 (1) | −0.001 (1) |
| C2 | 0.031 (1) | 0.032 (1) | 0.016 (1) | 0.001 (1) | −0.006 (1) | −0.001 (1) |
| C3 | 0.027 (1) | 0.035 (1) | 0.021 (1) | −0.005 (1) | −0.005 (1) | −0.002 (1) |
| C4 | 0.030 (1) | 0.036 (1) | 0.019 (1) | 0.000 (1) | −0.002 (1) | −0.001 (1) |
| C5 | 0.041 (2) | 0.058 (2) | 0.018 (1) | −0.012 (1) | −0.008 (1) | −0.004 (1) |
| C6 | 0.044 (2) | 0.072 (2) | 0.030 (2) | −0.029 (2) | −0.006 (1) | −0.002 (1) |
| C7 | 0.041 (2) | 0.056 (2) | 0.021 (1) | −0.016 (1) | 0.000 (1) | 0.002 (1) |
| C8 | 0.036 (2) | 0.038 (2) | 0.056 (2) | −0.003 (1) | −0.013 (1) | 0.007 (1) |
| C9 | 0.044 (2) | 0.036 (2) | 0.053 (2) | 0.006 (1) | −0.012 (1) | 0.009 (1) |
| C10 | 0.035 (1) | 0.034 (1) | 0.049 (2) | 0.000 (1) | −0.004 (1) | 0.006 (1) |
Geometric parameters (Å, º) top
| Cu1—N1 | 1.978 (2) | C2—C3 | 1.396 (3) |
| Cu1—N1i | 1.978 (2) | C3—C4 | 1.387 (3) |
| Cu1—O1 | 1.980 (2) | C4—C5 | 1.384 (4) |
| Cu1—O1i | 1.980 (2) | C5—C6 | 1.386 (4) |
| Cu1—O3ii | 2.646 (2) | C6—C7 | 1.377 (4) |
| Cu1—O3iii | 2.646 (2) | C8—C9 | 1.354 (4) |
| O1—C1 | 1.284 (3) | O3—H3o | 0.85 (1) |
| O2—C1 | 1.239 (3) | N2—H2n | 0.85 (1) |
| O3—C4 | 1.366 (3) | C3—H3 | 0.93 |
| N1—C10 | 1.310 (4) | C5—H5 | 0.93 |
| N1—C8 | 1.376 (4) | C6—H6 | 0.93 |
| N2—C10 | 1.338 (4) | C7—H7 | 0.93 |
| N2—C9 | 1.357 (4) | C8—H8 | 0.93 |
| C1—C2 | 1.500 (3) | C9—H9 | 0.93 |
| C2—C7 | 1.388 (4) | C10—H10 | 0.93 |
| | | |
| N1—Cu1—N1i | 180 | O3—C4—C5 | 121.5 (2) |
| N1—Cu1—O1 | 89.0 (1) | O3—C4—C3 | 118.5 (2) |
| N1—Cu1—O1i | 91.1 (1) | C5—C4—C3 | 119.9 (2) |
| N1—Cu1—O3ii | 88.52 (1) | C4—C5—C6 | 119.7 (2) |
| N1—Cu1—O3iii | 91.5 (1) | C7—C6—C5 | 120.8 (3) |
| N1i—Cu1—O1 | 91.1 (1) | C6—C7—C2 | 119.7 (2) |
| N1i—Cu1—O1i | 89.0 (1) | C9—C8—N1 | 109.8 (3) |
| O1—Cu1—O1i | 180 | C8—C9—N2 | 105.8 (3) |
| N1i—Cu1—O3iii | 88.52 (8) | N1—C10—N2 | 111.6 (3) |
| O1—Cu1—O3iii | 86.55 (7) | C4—O3—H3o | 109 (3) |
| O1i—Cu1—O3iii | 93.45 (7) | C10—N2—H2n | 123 (3) |
| N1i—Cu1—O3ii | 91.48 (8) | C9—N2—H2n | 129 (3) |
| O1—Cu1—O3ii | 93.45 (7) | C4—C3—H3 | 120.0 |
| O1i—Cu1—O3ii | 86.55 (7) | C2—C3—H3 | 120.0 |
| O3iii—Cu1—O3ii | 180.0 | C4—C5—H5 | 120.1 |
| C1—O1—Cu1 | 127.5 (2) | C6—C5—H5 | 120.1 |
| C10—N1—C8 | 105.1 (2) | C7—C6—H6 | 119.6 |
| C10—N1—Cu1 | 126.5 (2) | C5—C6—H6 | 119.6 |
| C8—N1—Cu1 | 127.9 (2) | C6—C7—H7 | 120.1 |
| C10—N2—C9 | 107.7 (2) | C2—C7—H7 | 120.1 |
| O2—C1—O1 | 124.9 (2) | C9—C8—H8 | 125.1 |
| O2—C1—C2 | 119.1 (2) | N1—C8—H8 | 125.1 |
| O1—C1—C2 | 115.9 (2) | C8—C9—H9 | 127.1 |
| C7—C2—C3 | 119.7 (2) | N2—C9—H9 | 127.1 |
| C7—C2—C1 | 119.9 (2) | N1—C10—H10 | 124.2 |
| C3—C2—C1 | 120.4 (2) | N2—C10—H10 | 124.2 |
| C4—C3—C2 | 120.0 (2) | | |
| | | |
| N1i—Cu1—O1—C1 | −97.3 (2) | C7—C2—C3—C4 | −1.0 (4) |
| N1—Cu1—O1—C1 | 82.7 (2) | C1—C2—C3—C4 | 176.7 (2) |
| O3iii—Cu1—O1—C1 | 174.3 (2) | C2—C3—C4—O3 | −175.1 (2) |
| O3ii—Cu1—O1—C1 | −5.7 (2) | C2—C3—C4—C5 | 2.4 (4) |
| O1—Cu1—N1—C10 | 122.8 (3) | O3—C4—C5—C6 | 176.0 (3) |
| O1i—Cu1—N1—C10 | −57.2 (3) | C3—C4—C5—C6 | −1.5 (4) |
| O3iii—Cu1—N1—C10 | 36.3 (3) | C4—C5—C6—C7 | −0.9 (5) |
| O3ii—Cu1—N1—C10 | −143.7 (3) | C5—C6—C7—C2 | 2.3 (5) |
| O1—Cu1—N1—C8 | −48.2 (3) | C3—C2—C7—C6 | −1.3 (4) |
| O1i—Cu1—N1—C8 | 131.8 (3) | C1—C2—C7—C6 | −179.0 (3) |
| O3iii—Cu1—N1—C8 | −134.7 (3) | C10—N1—C8—C9 | −0.7 (4) |
| O3ii—Cu1—N1—C8 | 45.3 (3) | Cu1—N1—C8—C9 | 171.8 (2) |
| Cu1—O1—C1—O2 | 9.4 (4) | N1—C8—C9—N2 | 0.7 (4) |
| Cu1—O1—C1—C2 | −170.1 (2) | C10—N2—C9—C8 | −0.4 (4) |
| O2—C1—C2—C7 | 25.4 (4) | C8—N1—C10—N2 | 0.5 (4) |
| O1—C1—C2—C7 | −155.1 (3) | Cu1—N1—C10—N2 | −172.2 (2) |
| O2—C1—C2—C3 | −152.3 (3) | C9—N2—C10—N1 | −0.1 (4) |
| O1—C1—C2—C3 | 27.2 (3) | | |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y, z−1; (iii) −x+1, −y+1, −z+2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3o···O2iv | 0.85 (1) | 1.76 (1) | 2.596 (3) | 170 (4) |
| N2—H2n···O1v | 0.85 (1) | 2.01 (2) | 2.841 (3) | 166 (4) |
| Symmetry codes: (iv) x, y, z+1; (v) −x+1, y−1/2, −z+3/2. |
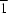 . Above and below the square are the hydroxy O atoms of adjacent molecules. This weak interaction [2.646 (2) Å] leads to the formation of a linear chain; the chains are consolidated into a layer through hydrogen bonds.
. Above and below the square are the hydroxy O atoms of adjacent molecules. This weak interaction [2.646 (2) Å] leads to the formation of a linear chain; the chains are consolidated into a layer through hydrogen bonds.![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.004 Å
(C-C) = 0.004 ÅAlert level C PLAT094_ALERT_2_C Ratio of Maximum / Minimum Residual Density .... 2.22 PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra O3 -H3O 1.555 1.555 PLAT731_ALERT_1_C Bond Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra N2 -H2N 1.555 1.555 PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra O3 -H3O 1.555 1.555 PLAT735_ALERT_1_C D-H Calc 0.85(3), Rep 0.850(10) ...... 3.00 su-Ra N2 -H2N 1.555 1.555 PLAT736_ALERT_1_C H...A Calc 1.76(3), Rep 1.760(10) ...... 3.00 su-Ra H3O -O2 1.555 1.556

 journal menu
journal menu










 Alert level C
Alert level C



