issue contents
September 2010 issue
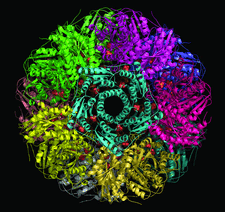
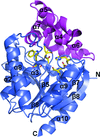
Cover illustration: Icosahedral assembly of lumazine synthase from Bacillus anthracis (p. 1001). The view is along the fivefold icosahedral axis. Each pentamer is shown in a different colour and inorganic phosphate ions found in the active sites are shown as spheres.
research papers
The structures of the B. subtilis genomic dUTPase and of its complex with the substrate analogue dUpNHpp and calcium are reported, both at 1.85 Å resolution. The Phe-lid, which folds over the ligand, lies in a novel position in the structure.
The structures of three DhaA mutants at atomic resolution are described in detail and compared in order to explore the effect of mutations on the enzymatic activity of modified proteins from a structural perspective.
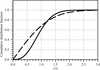
A new percentile-based measure of structural differences is proposed and is shown to have advantages over traditional root-mean-square distance.
The crystal structures of the nucleotide-binding domain of human ABCB6 in the apo form and in complexes with ADP, with ATP and with ADP and Mg2+ were determined at 2 Å resolution.
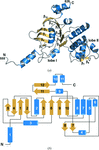
New insights into group II intron self-splicing mechanisms from an improved crystal structure of a group II intron complex with a product upon inclusion of weak high-resolution X-ray in structure determination.
PDB reference: group II intron, 3g78
Open  access
access
 access
accessCrystallographic studies of lumazine synthase, the penultimate enzyme of the riboflavin-biosynthetic pathway in B. anthracis, provide a structural framework for the design of antibiotic inhibitors, together with calorimetric and kinetic investigations of inhibitor binding.
Main-chain tripeptidic and tetrapeptidic molecular fragments are positioned in the electron density by a phased rotation, conformation and translation function at various resolutions. Accurate and nearly complete protein models can be built semi-automatically using the positioned fragments.
Open  access
access
 access
accessA new method for modeling the bulk solvent in macromolecular diffraction data based on Babinet's principle is presented. The proposed models offer the advantage of differentiability with respect to atomic coordinates.
short communications
Open  access
access
 access
accessA grid-scan tool that enables rapid characterization of large sample volumes using a microfocused X-ray beam and a fast-readout detector is reported.
Open  access
access
 access
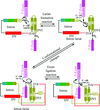
accessThe glycolytic enzyme enolase associates with the endoribonuclease RNase E in Escherichia coli and many other bacterial species. The crystal structure of the complex reveals the basis for the molecular recognition and provides clues as to the possible function of the interaction.
PDB reference: enolase–RNase E recognition domain complex, 3h8a
letters to the editor
Open  access
access
 access
accessA brief comment is made on the need to use carefully selected, novel terms in crystallographic publications, especially publications addressing non-specialists.


 journal menu
journal menu