issue contents
November 2010 issue
Neutrons in biology
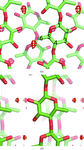
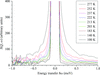
Cover illustration: (Fcalc, ![[varphi]](/logos/entities/phiv_rmgif.gif) calc) Fourier syntheses computed at different resolutions (top to bottom: 1, 1.5, 2 and 2.5 Å) and corresponding to four different cases (left to right): X-ray, neutron fully deuterated, neutron fully hydrogenated and neutron partially deuterated, in which the hydroxyl H (HH) of tyrosine shares its site with deuterium (DH) with an occupancy ratio of 0.6:0.4 (p. 1153).
calc) Fourier syntheses computed at different resolutions (top to bottom: 1, 1.5, 2 and 2.5 Å) and corresponding to four different cases (left to right): X-ray, neutron fully deuterated, neutron fully hydrogenated and neutron partially deuterated, in which the hydroxyl H (HH) of tyrosine shares its site with deuterium (DH) with an occupancy ratio of 0.6:0.4 (p. 1153).
editorial
Free 

A summary of the current state of neutron crystallography is given.
introduction
A summary of the International Conference on Neutrons in Biology is presented.
research papers
The crystal structures of drug-target proteins, such as human immunodeficiency virus protease and pancreatic elastase, in complex with transition-state analogue inhibitors have been determined by neutron diffraction.
The structure and mechanism of diisopropyl fluorophosphatase (DFPase) have been studied using a variety of methods, including isotopic labelling, X-ray crystallography and neutron crystallography. The neutron structure of DFPase, mechanistic studies and subsequent rational design efforts are described.
Neutron crystallography can make an important contribution to our understanding of the structural basis of sweetness, as illustrated by results on studies of the sweet protein thaumatin.
Using neutron diffraction analysis, the protonation states of 35 of 38 histidine residues were determined for the deoxy form of normal human adult hemoglobin. Distal and buried histidines may contribute to the increased affinity of the deoxy state for hydrogen ions and its decreased affinity for oxygen compared with the oxygenated form.
The implementation of crystallographic structure-refinement procedures that include both X-ray and neutron data (separate or jointly) in the PHENIX system is described.
Two new computational tools have been developed for H/D determination in macromolecular structures from neutron data.
A series of cellulose crystal allomorphs have been studied using high-resolution X-ray and neutron fibre diffraction to locate the positions of H atoms involved in hydrogen bonding.
The first neutron crystal structure of carbonic anhydrase is presented. The structure reveals interesting and unexpected features of the active site that affect catalysis.
The current state of in silico studies on cellulose is reviewed and commented on.
The multi-scale structures of cellulose extracted from native and dilute acid pretreated switchgrass, as well as that of microcrystalline cellulose (Avicel), were investigated using SANS.
A new diffractometer (iBIX) at the next-generation neutron source at J-PARC has begun operation for protein crystallography. Preliminary structure analyses of organic crystals demonstrate that iBIX has high performance even at 120 kW operation.
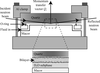
The LADI-III instrument for measuring neutron macromolecular crystallography diffraction data is described. The data collection of type-III perdeuterated antifreeze protein is given as an example.
The Protein Crystallography Station user facility at Los Alamos National Laboratory not only offers open access to a high-performance neutron beamline, but also actively supports and develops new methods in protein expression, deuteration, purification, robotic crystallization and the synthesis of substrates with stable isotopes and provides assistance with data-reduction and structure-refinement software and comprehensive neutron structure analysis.
Small-angle neutron scattering with contrast variation is a powerful tool for probing complex biological macromolecular complexes.
The binding of microtubules by the molecular motor Ncd in solution was investigated using small-angle neutron scattering. In the presence of an ATP analogue one of the two Ncd motor heads is cooperatively bound to a tubulin dimer.
This article is a plea to neutron scatterers studying protein dynamics to move on from measuring a few model systems and arguing about the physics in order to address the diversity and specificity of proteins and provide much-needed data for the understanding of the essential role of dynamics in biological function and activity.
Neutron scattering experiments with myoglobin over extended time and temperature ranges provide insight into protein dynamics.
Neutron diffraction from hydrated stacks of natural two-dimensional crystal patches of purple membrane from Halobacterium salinarum was studied as a function of pressure.
In this article, methods are presented for the creation and characterization of supported model membranes which can mimic many of the critical attributes of cell membranes. It is demonstrated that neutron reflectometry can characterize the structure, composition and organization of model membranes deposited on solid, nanoporous and polymer supports.
Neutron diffraction methods can be used to provide novel information on the structure and behaviour of nucleic acids as studied in single crystals, fibres and solutions.
X-ray and neutron diffraction studies of cyanomethemoglobin are being used to evaluate the structural waters within the dimer–dimer interface involved in quaternary-state transitions.
A description is given of the results of neutron diffraction studies of the structures of four different metal-ion complexes of deuterated D-xylose isomerase.
A history of neutrons in biology: the development of neutron protein crystallography at BNL and LANL
Neutron scattering techniques provide valuable information on the structure of biological molecules and molecular systems, and hence provide insight into function.


 journal menu
journal menu