issue contents
May 2016 issue

Cover illustration: Negative-stained transmission electron microscopy image of RNA polymerase II (Pol II) crystal fragments illustrating high quality crystal lattices and Bragg spots (inset) (Stevenson et al., p. 603). Highly contrasted individual particles of Pol II (background) can be seen detaching from the upper plane of the larger crystal fragment.
scientific commentaries
Systematic analysis of a protein–RNA complex has shown how nucleoprotein complexes can be protected from radiation damage.
research papers
In this article, the potential of transmission electron microscopy to assist in the process of generating well diffracting crystals for conventional crystallography, as well as for free-electron laser and micro-electron diffraction applications, is demonstrated.
An automated pipeline, I-TASSER-MR, has been developed to facilitate structure determination using molecular repacement (MR) for samples for which only distantly related structures are available in the Protein Data Bank. By combining iterative fragmental structure-assembly simulations and progressive trimming of poorly modeled regions, the pipeline improves the success rate of MR.
Open  access
access
 access
accessA generic protocol for investigating crystal dehydration is presented and tested with a set of protein crystal systems using the HC1b high-precision crystal humidifier/dehumidifier.
The structures of E. coli branching enzyme bound to α-, β- and γ-cyclodextrin reveal a novel substrate-binding site and illuminate the determinants of glucan binding specificity.
Open  access
access
 access
accessSystematic analysis of radiation damage within a protein–RNA complex over a large dose range (1.3–25 MGy) reveals significant differential susceptibility of RNA and protein. A new method of difference electron-density quantification is presented.
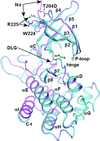
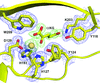
To find a crystallization-competent TGFβR2 kinase domain, several surface patches of potentially high-entropy residues were identified. Constructs with these residues mutated to alanine were expressed and purified. One yielded crystals that routinely diffracted to better than 2 Å resolution. Comparison of structures of TGFβR1 and TGFβR2 with inhibitors explain the isoform selectivity.
PDB references: apo TGFβR1-WT, 5e8s; apo TGFβR1-T204D, 5e8t; apo TGFβR1-8M, 5e8u; apo TGFβR2-6M, 5e8v; TGFβR1-T204D, complex with staurosporine, 5e8w; TGFβR1-8M, complex with staurosporine, 5e8x; TGFβR2-6M, complex with staurosporine, 5e8y; TGFβR1-T204D, complex with compound 1, 5e8z; TGFβR1-8M, complex with compound 1, 5e90; TGFβR2-6M, complex with compound 1, 5e91; TGFβR2-6M, complex with AMPPNP, 5e92
Active-site reorganization of a prolyl 4-hydroxylase from B. anthracis is observed upon the binding of metal and α-ketoglutarate.
Open  access
access
 access
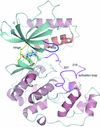
accessCrystal structures of the MAP kinase ERK5 in complex with XMD8-92 and four novel inhibitors reveal an allosteric binding site between the kinase P-loop and αC helix. Binding at this site displaces the P-loop into the ATP-binding site and was shown to be ATP-competitive.
A K354E mutation in tomato leucine aminopeptidase shifts the oligomerization equilibrium towards lower-order oligomers, exposing hydrophobic surfaces that are likely to be involved in chaperone activity.
PDB reference: tomato leucine aminopeptidase, K354E mutant, 5d8n


 journal menu
journal menu