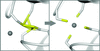issue contents
October 2016 issue

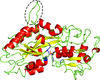
Cover illustration: The formylation-tailoring and adenylation domains of the initiation module of linear gramicidin synthetase (F-A-PCP) have undergone manipulation to produce a new crystal form which has an unanticipated packing (Reimer et al., p. 1130). This figure shows the pseudo-independent packing networks in a new P41 crystal. Molecules A and C (in green) form a packing network essentially identical to that of an unmodified crystal, and molecules B and D (in magenta and purple) form a separate, translationally related network, and these combine to result in the F-Asub lattice.
research papers
The structure of a serendipitously crystallized protein was determined using a large-scale molecular replacement protocol, and on the basis of the sequence deduced from the electron-density map the protein was identified as a phosphate-binding protein from S. maltophilia.
Open  access
access
 access
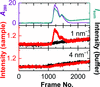
accessSAXS coupled with online ion-exchange chromatography allows the collection of high-quality BioSAXS data.
High-resolution structures of the cytosolic disulfide reductase DsbM in apo and S-glutathionylated states are described.
Open  access
access
 access
accessA method is presented to automatically validate and correct Zn–CysxHisy complexes that have a distorted tetrahedral geometry.
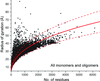
Power laws describing the dependence of the radius of gyration on the number of residues are calculated for protein oligomers. The power laws are useful for predicting the oligomeric state from small-angle X-ray scattering, identifying elongated proteins and validating the annotation of biological assemblies.
Open  access
access
 access
accessA nonribosomal peptide synthetase di-domain construct was produced using known crystal packing as a guide, and the resulting crystal has an unanticipated packing.
A new approach is reported to computationally generate plausible structural models using a procedure that combines crystallographic structures and density maps obtained from three-dimensional electron microscopy.
Open  access
access
 access
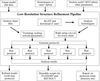
accessAn automated pipeline for low-resolution structure refinement (LORESTR) has been developed to assist in the hassle-free refinement of difficult cases. The pipeline automates the selection of high-resolution homologues for external restraint generation and optimizes the parameters for ProSMART and REFMAC5, improving R factors and geometry statistics in 94% of the test cases.


 journal menu
journal menu