issue contents
October 2017 issue
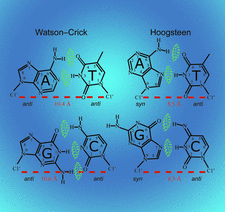
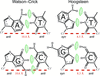
Cover illustration: Watson-Crick and Hoogsteen base-pair conformations seen in DNA (Hintze et al., p. 852).
research papers
Download citation


Download citation


Open  access
access
 access
accessThe different conformations of WalK reveal the intrinsic dynamic properties of a sensor histidine kinase structure as a molecular basis for multifunctionality.
Open  access
access
 access
accessNanobodies are used as crystallization chaperones and here site-specific mercury labelling of nanobodies is shown as a new tool for phasing.
1.8 Å resolution crystal structures of a Francisella protein required for growth in host cells are reported.
Heavily glycosylated structures of an HIV-1 envelope trimer derived by X-ray crystallography and cryo-EM are compared.
Open  access
access
 access
accessNew compact and precise cryogenic sample holders for macromolecular crystallography are proposed as possible future European standards.
Open  access
access
 access
accessA sample changer based on a six-axis industrial robot and a new sample-storage dewar with an ice-cleaning feature have been developed to open automated X-ray crystallography beamlines to new sample-holder models, such as miniSPINE and NewPin, while remaining compatible with the widespread SPINE sample-holder standard.
This paper describes a method that employs difference electron density to identify DNA purines modeled in the incorrect syn/anti conformation. Different classes of mistakes that were identified by using this method to survey all PDB models containing DNA are highlighted.


 journal menu
journal menu