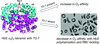issue contents
November 2018 issue
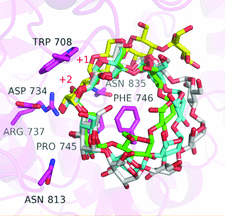
Cover illustration: Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin (Saka et al., p.1115). Shown here is the interaction mode in the active site where the magenta, yellow, green, cyan and grey sticks are the binding residues for maltotetraose in the KPP-maltotetraose complex structure, ![[alpha]](/logos/entities/alpha_rmgif.gif) -cyclodextrin,
-cyclodextrin, ![[beta]](/logos/entities/beta_rmgif.gif) -cyclodextrin and
-cyclodextrin and ![[gamma]](/logos/entities/gamma_rmgif.gif) -cyclodextrin, respectively.
-cyclodextrin, respectively.
scientific commentaries
Highlighting the structure-based approach to combatting sickle cell disease taken by Safo and coworkers.
research papers
Open  access
access
 access
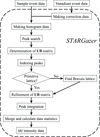
accessIn this article, the status of the STARGazer data-processing software and its data-processing algorithms are described in detail. The STARGazer data-processing software is used for neutron time-of-flight single-crystal diffraction data collected using the IBARAKI Biological Crystal Diffractometer.
The crystal structures of two fimbrial adhesins from the uropathogen P. mirabilis are reported as a first step towards unveiling their role in pathogenesis. Despite lacking significant sequence homology, the two structures are very similar.
Open  access
access
 access
accessQuantum-mechanics/molecular-mechanics (ONIOM) X-ray macromolecular refinement using the program DivCon integrated with PHENIX is reported.
The crystal structure of α-L-rhamnosidase, a biotechnologically important enzyme isolated from Aspergillus terreus, reveals its active-site architecture, domain organization and native glycosylation.
PDB reference: rhamnosidase from A. terreus, 6gsz
Open  access
access
 access
accessIt is demonstrated that using three-dimensional profile fitting of Bragg peaks increases the accuracy and resolution of neutron crystallographic data collected from proteins and reveals new features in nuclear density maps calculated from these data.
The program Buccaneer has been integrated in a cyclic automatic model-building procedure. improving its efficiency and accuracy.
A detailed analysis of a winged helix–turn–helix family transcriptional regulator that positively regulates archaeal biofilm formation is reported.
PDB reference: archaeal biofilm regulator, 6cmv
Crystal structures of Klebsiella pneumoniae pullulanase (KPP) in complex with α-, β- or γ-cyclodextrin were determined at around 1.98–2.59 Å resolution. The cyclodextrins interact with CBM41 and the active site of KPP. In the active site, Phe746 interacts with the inner cavity of the cyclodextrin rings. The strong inhibition of KPP by β-cyclodextrin is caused by the optimized interaction between β-cyclodextrin and the side chain of Phe746. These structures and the kinetic parameters of recombinant pullulanase and its F746A variant revealed the importance of Phe746 in the selective binding of cyclodextrins.
PDB references: Klebsiella pneumoniae pullulanase, apo, space group P43212, 5yn2; complex formed in the presence of 0.1 mM β-cyclodextrin, 5yn7; complex formed in the presence of 1 mM α-cyclodextrin, 5yna; complex formed in the presence of 1 mM β-cyclodextrin, 5ync; complex formed in the presence of 1 mM γ-cyclodextrin, 5ynd; complex formed in the presence of 10 mM α-cyclodextrin, 5yne; complex formed in the presence of 10 mM γ-cyclodextrin, 5ynh
book reviews
Free 



 journal menu
journal menu