

metal-organic compounds
Di-μ-chlorido-bis[(2,2′:6′,2′′-terpyridine-κ3N,N′,N′′)copper(II)] bis(trifluoromethanesulfonate)
aDepartment of Chemistry and Biochemistry, University of the Incarnate Word, San Antonio TX 78209, USA, and bDepartment of Chemistry, The University of Texas at San Antonio, San Antonio TX 78249, USA
*Correspondence e-mail: adrian@uiwtx.edu
In the centrosymmetric title complex, [Cu2Cl2(C15H11N3)2](CF3O3S)2, the CuII metal center is fivefold coordinated by two chloride ions and three nitrogen atoms of the terpyridine ligand in a distorted square-pyramidal geometry; two trifluoromethanesulfonate ions complete the outer coordination sphere. π–π stacking interactions between the pyridyl rings in adjacent molecules contribute to the alignment of the complexes in columns along the a-axis. This structure represents the first example of a binuclear dication of formula [Cu(terpy)2Cl2]2+ with trifluoromethanesulfonate as counter-ions.
Keywords: crystal structure; terpyridine; copper; trifluoromethanesulfonate salt; bridging chloride; π–π stacking.
CCDC reference: 2116881
![[Scheme 3D1]](bt4119scheme3D1.gif)
![[Scheme 1]](bt4119scheme1.gif)
Structure description
Terpyridines are some of the most studied nitrogen-based tridentate ligands in coordination chemistry, and their metal complexes have found application in catalysis (Wei et al., 2019![Wei, C., He, Y., Shi, X. & Song, Z. (2019). Coord. Chem. Rev. 385, 1-19. [Wei, C., He, Y., Shi, X. & Song, Z. (2019). Coord. Chem. Rev. 385, 1-19.]](../../../../../../logos/arrows/iucrx_arr.gif) ; Choroba et al., 2019
; Choroba et al., 2019![Choroba, K., Machura, B., Kula, S., Raposo, L. R., Fernandes, A. R., Kruszynski, R., Erfurt, K., Shul'pina, L. S., Kozlov, Y. N. & Shul'pin, G. B. (2019). Dalton Trans. 48, 12656-12673. [Choroba, K., Machura, B., Kula, S., Raposo, L. R., Fernandes, A. R., Kruszynski, R., Erfurt, K., Shul'pina, L. S., Kozlov, Y. N. & Shul'pin, G. B. (2019). Dalton Trans. 48, 12656-12673.]](../../../../../../logos/arrows/iucrx_arr.gif) ), supramolecular chemistry (Wei et al., 2019
), supramolecular chemistry (Wei et al., 2019![Wei, C., He, Y., Shi, X. & Song, Z. (2019). Coord. Chem. Rev. 385, 1-19. [Wei, C., He, Y., Shi, X. & Song, Z. (2019). Coord. Chem. Rev. 385, 1-19.]](../../../../../../logos/arrows/iucrx_arr.gif) ), and medicinal chemistry (Glišić et al., 2018
), and medicinal chemistry (Glišić et al., 2018![Glišić, B. Đ., Nikodinovic-Runic, J., Ilic-Tomic, T., Wadepohl, H., Veselinović, A., Opsenica, I. M. & Djuran, M. I. (2018). Polyhedron, 139, 313-322. [Glišić, B. Đ., Nikodinovic-Runic, J., Ilic-Tomic, T., Wadepohl, H., Veselinović, A., Opsenica, I. M. & Djuran, M. I. (2018). Polyhedron, 139, 313-322.]](../../../../../../logos/arrows/iucrx_arr.gif) ; Malarz et al., 2021
; Malarz et al., 2021![Malarz, K., Zych, D., Gawecki, R., Kuczak, M., Musioł, R. & Mrozek-Wilczkiewicz, A. (2021). Eur. J. Med. Chem. 212, 113032. [Malarz, K., Zych, D., Gawecki, R., Kuczak, M., Musioł, R. & Mrozek-Wilczkiewicz, A. (2021). Eur. J. Med. Chem. 212, 113032.]](../../../../../../logos/arrows/iucrx_arr.gif) ; Li et al., 2020
; Li et al., 2020![Li, C., Xu, F., Zhao, Y., Zheng, W., Zeng, W., Luo, Q., Wang, Z., Wu, K., Du, J. & Wang, F. (2020). Front. Chem. 8, 210. [Li, C., Xu, F., Zhao, Y., Zheng, W., Zeng, W., Luo, Q., Wang, Z., Wu, K., Du, J. & Wang, F. (2020). Front. Chem. 8, 210.]](../../../../../../logos/arrows/iucrx_arr.gif) ). Recently, copper(II) terpyridine complexes have received much attention due to their remarkable cytotoxicity and ability to interact with DNA (Karges et al., 2021
). Recently, copper(II) terpyridine complexes have received much attention due to their remarkable cytotoxicity and ability to interact with DNA (Karges et al., 2021![Karges, J., Xiong, K., Blacque, O., Chao, H. & Gasser, G. (2021). Inorg. Chim. Acta, 516, 120137. [Karges, J., Xiong, K., Blacque, O., Chao, H. & Gasser, G. (2021). Inorg. Chim. Acta, 516, 120137.]](../../../../../../logos/arrows/iucrx_arr.gif) ); herein, we report the synthesis and structure of the title copper(II) terpyridine complex.
); herein, we report the synthesis and structure of the title copper(II) terpyridine complex.
The of the title compound, depicted in Fig. 1![[link]](../../../../../../logos/arrows/iucrx_arr.gif) , consists of half of a centrosymmetric dication [Cu(terpy)2Cl2]2+ and one trifluoromethanesulfonate ion completing the outer coordination sphere. The Cu—N, and Cu—Cl distances, as well as, the Cl—Cu—Cl, N—Cu—Cl and N—Cu—N angles are in good agreement with the reported values in similar copper(II) terpyridine complexes currently available in the CSD (version 5.42 with update September 2021; Rojo et al., 1987
, consists of half of a centrosymmetric dication [Cu(terpy)2Cl2]2+ and one trifluoromethanesulfonate ion completing the outer coordination sphere. The Cu—N, and Cu—Cl distances, as well as, the Cl—Cu—Cl, N—Cu—Cl and N—Cu—N angles are in good agreement with the reported values in similar copper(II) terpyridine complexes currently available in the CSD (version 5.42 with update September 2021; Rojo et al., 1987![Rojo, T., Arriortua, M. I., Ruiz, J., Darriet, J., Villeneuve, G. & Beltran-Porter, D. (1987). J. Chem. Soc. Dalton Trans. pp. 285-291. [Rojo, T., Arriortua, M. I., Ruiz, J., Darriet, J., Villeneuve, G. & Beltran-Porter, D. (1987). J. Chem. Soc. Dalton Trans. pp. 285-291.]](../../../../../../logos/arrows/iucrx_arr.gif) ; refcode FECJEC; Valdés-Martínez et al., 2002;
; refcode FECJEC; Valdés-Martínez et al., 2002;![Valdés-Martínez, J., Salazar-Mendoza, D. & Toscano, R. A. (2002). Acta Cryst. E58, m712-m714. [Valdés-Martínez, J., Salazar-Mendoza, D. & Toscano, R. A. (2002). Acta Cryst. E58, m712-m714.]](../../../../../../logos/arrows/iucrx_arr.gif) refcode HULZAP; Gasser et al., 2004
refcode HULZAP; Gasser et al., 2004![Gasser, G., Labat, G. & Stoeckli-Evans, H. (2004). Acta Cryst. E60, m244-m246. [Gasser, G., Labat, G. & Stoeckli-Evans, H. (2004). Acta Cryst. E60, m244-m246.]](../../../../../../logos/arrows/iucrx_arr.gif) ; refcode HULZAP01). All relevant bond lengths and angles involving the Cu atom are presented in Table 1
; refcode HULZAP01). All relevant bond lengths and angles involving the Cu atom are presented in Table 1![[link]](../../../../../../logos/arrows/iucrx_arr.gif) .
.
|
![[Figure 1]](bt4119fig1thm.gif) | Figure 1 The molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level; H atoms are omitted for clarity. Symmetry operator for generating equivalent atoms: (i) 1 − x, 1 − y, 1 − z. |
In the crystal packing of the title compound, π–π stacking interactions between the N1 and N3 pyridyl ring of adjacent molecules are observed, with a centroid-to-centroid (Cg⋯Cg) distance of 3.658 (1) Å and an offset distance of 1.723 Å. No other supramolecuar interaction is present in the crystal packing of the title compound.
Synthesis and crystallization
The title compound was obtained as product of the reaction of 2,2′:6′,2′′-terpyridine (0.100 g, 0.429 mmol) with copper(II) chloride dihydrate (0.073 g, 0.429 mmol) in acetonitrile after the addition of silver trifluoromethanesulfonate (0.110 g, 0.429 mmol) and filtration using a 0.45 µm PTFE syringe filter. Crystals suitable for X-ray diffraction of the title compound were obtained by vapor diffusion of diethyl ether over the resulting acetonitrile solution at 278 K.
Refinement
Crystal data, data collection and structure details are summarized in Table 2![[link]](../../../../../../logos/arrows/iucrx_arr.gif) . H atoms were located in a difference map and refined in idealized positions using a riding model with atomic displacement parameters of Uiso(H) = 1.2Ueq(C) and with a C—H distance of 0.95 Å.
. H atoms were located in a difference map and refined in idealized positions using a riding model with atomic displacement parameters of Uiso(H) = 1.2Ueq(C) and with a C—H distance of 0.95 Å.
|
Structural data
CCDC reference: 2116881
contains datablock I. DOI: https://doi.org/10.1107/S2414314621010968/bt4119sup1.cif
Structure factors: contains datablock I. DOI: https://doi.org/10.1107/S2414314621010968/bt4119Isup2.hkl
Supporting information file. DOI: https://doi.org/10.1107/S2414314621010968/bt4119Isup3.mol
Data collection: CrysAlis PRO (Rigaku OD, 2019); cell CrysAlis PRO (Rigaku OD, 2019); data reduction: CrysAlis PRO (Rigaku OD, 2019); program(s) used to solve structure: ShelXT (Sheldrick, 2015a); program(s) used to refine structure: SHELXL2018/3 (Sheldrick, 2015b); molecular graphics: OLEX2 (Dolomanov et al., 2009); software used to prepare material for publication: OLEX2 (Dolomanov et al., 2009).
| [Cu2Cl2(C15H11N3)2](CF3O3S)2 | Z = 1 |
| Mr = 962.65 | F(000) = 482 |
| Triclinic, P1 | Dx = 1.846 Mg m−3 |
| a = 7.2767 (2) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.8394 (2) Å | Cell parameters from 14071 reflections |
| c = 13.1746 (3) Å | θ = 3.1–29.5° |
| α = 106.667 (2)° | µ = 1.59 mm−1 |
| β = 91.226 (2)° | T = 98 K |
| γ = 105.453 (2)° | Block, clear bluish green |
| V = 866.08 (4) Å3 | 0.47 × 0.17 × 0.1 mm |
| XtaLAB AFC12 (RCD3): Kappa single diffractometer | 3889 reflections with I > 2σ(I) |
| Radiation source: Rotating-anode X-ray tube, Rigaku (Mo) X-ray Source | Rint = 0.047 |
| ω scans | θmax = 27.5°, θmin = 2.3° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2019) | h = −9→9 |
| Tmin = 0.741, Tmax = 1.000 | k = −12→12 |
| 33748 measured reflections | l = −16→17 |
| 3982 independent reflections |
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.036 | H-atom parameters constrained |
| wR(F2) = 0.094 | w = 1/[σ2(Fo2) + (0.0529P)2 + 0.6752P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max = 0.002 |
| 3982 reflections | Δρmax = 0.55 e Å−3 |
| 253 parameters | Δρmin = −0.41 e Å−3 |
| 0 restraints |
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. H atoms were located in a difference map and refined in idealized positions using a riding model with atomic displacement parameters of Uiso(H) = 1.2Ueq(C) and with a C—H distance of 0.95 Å. |
| x | y | z | Uiso*/Ueq | ||
| Cu1 | 0.49265 (4) | 0.41355 (3) | 0.60172 (2) | 0.01772 (10) | |
| Cl1 | 0.62384 (8) | 0.36614 (6) | 0.44902 (4) | 0.02026 (13) | |
| S1 | 0.50126 (8) | 0.15121 (6) | 0.79698 (4) | 0.02142 (13) | |
| F2 | 0.7785 (2) | 0.02432 (18) | 0.78751 (14) | 0.0396 (4) | |
| F1 | 0.6642 (2) | 0.06787 (19) | 0.93864 (12) | 0.0395 (4) | |
| F3 | 0.5041 (2) | −0.11260 (17) | 0.80642 (16) | 0.0478 (4) | |
| O1 | 0.6469 (2) | 0.29199 (17) | 0.83604 (12) | 0.0239 (3) | |
| O3 | 0.3333 (2) | 0.1385 (2) | 0.85354 (15) | 0.0336 (4) | |
| O2 | 0.4661 (3) | 0.0941 (2) | 0.68265 (13) | 0.0360 (4) | |
| N3 | 0.2255 (3) | 0.27262 (19) | 0.55326 (14) | 0.0188 (4) | |
| N2 | 0.3802 (3) | 0.45334 (19) | 0.73563 (14) | 0.0168 (3) | |
| N1 | 0.7290 (3) | 0.55151 (19) | 0.70135 (14) | 0.0184 (4) | |
| C11 | 0.1030 (3) | 0.2851 (2) | 0.62992 (16) | 0.0192 (4) | |
| C5 | 0.6901 (3) | 0.6111 (2) | 0.80261 (16) | 0.0189 (4) | |
| C10 | 0.1958 (3) | 0.3840 (2) | 0.73629 (16) | 0.0186 (4) | |
| C6 | 0.4902 (3) | 0.5499 (2) | 0.82254 (16) | 0.0179 (4) | |
| C9 | 0.1110 (3) | 0.4087 (2) | 0.83124 (17) | 0.0220 (4) | |
| H9 | −0.019262 | 0.358669 | 0.833457 | 0.026* | |
| C15 | 0.1555 (3) | 0.1851 (2) | 0.45439 (17) | 0.0220 (4) | |
| H15 | 0.240897 | 0.174252 | 0.401071 | 0.026* | |
| C14 | −0.0386 (3) | 0.1094 (2) | 0.42706 (18) | 0.0238 (5) | |
| H14 | −0.084305 | 0.047636 | 0.356325 | 0.029* | |
| C12 | −0.0910 (3) | 0.2144 (2) | 0.60811 (17) | 0.0212 (4) | |
| H12 | −0.173957 | 0.225959 | 0.662670 | 0.025* | |
| C13 | −0.1631 (3) | 0.1253 (2) | 0.50390 (18) | 0.0231 (4) | |
| H13 | −0.296333 | 0.076425 | 0.486484 | 0.028* | |
| C7 | 0.4138 (3) | 0.5820 (2) | 0.91921 (16) | 0.0208 (4) | |
| H7 | 0.489630 | 0.651756 | 0.981351 | 0.025* | |
| C4 | 0.8284 (3) | 0.7197 (3) | 0.87846 (17) | 0.0225 (4) | |
| H4 | 0.798549 | 0.759683 | 0.948670 | 0.027* | |
| C8 | 0.2229 (3) | 0.5089 (3) | 0.92254 (17) | 0.0237 (5) | |
| H8 | 0.168203 | 0.527811 | 0.988179 | 0.028* | |
| C2 | 1.0514 (3) | 0.7057 (3) | 0.74756 (18) | 0.0241 (5) | |
| H2 | 1.176548 | 0.735707 | 0.727006 | 0.029* | |
| C16 | 0.6177 (3) | 0.0264 (3) | 0.83346 (19) | 0.0266 (5) | |
| C1 | 0.9066 (3) | 0.5979 (2) | 0.67581 (17) | 0.0216 (4) | |
| H1 | 0.934595 | 0.555208 | 0.605681 | 0.026* | |
| C3 | 1.0114 (3) | 0.7691 (3) | 0.84983 (18) | 0.0254 (5) | |
| H3 | 1.107711 | 0.845362 | 0.899683 | 0.030* |
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cu1 | 0.02295 (16) | 0.01815 (15) | 0.01013 (14) | 0.00641 (11) | 0.00275 (10) | 0.00076 (10) |
| Cl1 | 0.0282 (3) | 0.0213 (2) | 0.0128 (2) | 0.0121 (2) | 0.00591 (19) | 0.00269 (19) |
| S1 | 0.0221 (3) | 0.0223 (3) | 0.0156 (3) | 0.0060 (2) | −0.00080 (19) | −0.0002 (2) |
| F2 | 0.0382 (9) | 0.0427 (9) | 0.0461 (9) | 0.0246 (7) | 0.0131 (7) | 0.0132 (7) |
| F1 | 0.0508 (10) | 0.0437 (9) | 0.0241 (7) | 0.0128 (8) | −0.0061 (7) | 0.0120 (7) |
| F3 | 0.0458 (10) | 0.0206 (7) | 0.0667 (12) | 0.0001 (7) | −0.0117 (8) | 0.0074 (7) |
| O1 | 0.0284 (8) | 0.0199 (7) | 0.0201 (8) | 0.0056 (6) | 0.0030 (6) | 0.0021 (6) |
| O3 | 0.0230 (8) | 0.0371 (10) | 0.0371 (10) | 0.0075 (7) | 0.0075 (7) | 0.0066 (8) |
| O2 | 0.0400 (10) | 0.0429 (11) | 0.0171 (8) | 0.0111 (9) | −0.0078 (7) | −0.0016 (7) |
| N3 | 0.0262 (9) | 0.0162 (8) | 0.0132 (8) | 0.0065 (7) | 0.0020 (7) | 0.0028 (7) |
| N2 | 0.0215 (9) | 0.0152 (8) | 0.0130 (8) | 0.0063 (7) | 0.0008 (6) | 0.0021 (6) |
| N1 | 0.0239 (9) | 0.0182 (8) | 0.0133 (8) | 0.0075 (7) | 0.0024 (7) | 0.0038 (7) |
| C11 | 0.0269 (11) | 0.0159 (9) | 0.0149 (9) | 0.0073 (8) | 0.0010 (8) | 0.0037 (8) |
| C5 | 0.0248 (11) | 0.0190 (10) | 0.0141 (9) | 0.0087 (8) | 0.0032 (8) | 0.0043 (8) |
| C10 | 0.0237 (10) | 0.0167 (9) | 0.0157 (10) | 0.0073 (8) | 0.0014 (8) | 0.0039 (8) |
| C6 | 0.0234 (10) | 0.0152 (9) | 0.0144 (9) | 0.0058 (8) | 0.0014 (8) | 0.0030 (8) |
| C9 | 0.0227 (11) | 0.0225 (10) | 0.0186 (10) | 0.0053 (9) | 0.0042 (8) | 0.0039 (8) |
| C15 | 0.0332 (12) | 0.0176 (10) | 0.0144 (10) | 0.0080 (9) | 0.0016 (8) | 0.0031 (8) |
| C14 | 0.0362 (12) | 0.0149 (9) | 0.0167 (10) | 0.0056 (9) | −0.0045 (9) | 0.0011 (8) |
| C12 | 0.0247 (11) | 0.0182 (10) | 0.0205 (10) | 0.0065 (8) | 0.0017 (8) | 0.0054 (8) |
| C13 | 0.0259 (11) | 0.0158 (10) | 0.0246 (11) | 0.0040 (8) | −0.0045 (9) | 0.0039 (8) |
| C7 | 0.0256 (11) | 0.0210 (10) | 0.0122 (9) | 0.0061 (9) | 0.0010 (8) | 0.0000 (8) |
| C4 | 0.0261 (11) | 0.0241 (11) | 0.0157 (10) | 0.0088 (9) | 0.0031 (8) | 0.0020 (8) |
| C8 | 0.0282 (12) | 0.0277 (11) | 0.0134 (10) | 0.0087 (9) | 0.0055 (8) | 0.0025 (8) |
| C2 | 0.0215 (11) | 0.0285 (11) | 0.0233 (11) | 0.0074 (9) | 0.0034 (8) | 0.0093 (9) |
| C16 | 0.0308 (12) | 0.0197 (10) | 0.0240 (11) | 0.0049 (9) | −0.0022 (9) | 0.0010 (9) |
| C1 | 0.0266 (11) | 0.0250 (11) | 0.0166 (10) | 0.0114 (9) | 0.0055 (8) | 0.0075 (8) |
| C3 | 0.0256 (11) | 0.0275 (11) | 0.0198 (11) | 0.0063 (9) | −0.0014 (9) | 0.0036 (9) |
| Cu1—Cl1 | 2.2265 (5) | C10—C9 | 1.394 (3) |
| Cu1—Cl1i | 2.7660 (6) | C6—C7 | 1.387 (3) |
| Cu1—N3 | 2.0278 (19) | C9—H9 | 0.9500 |
| Cu1—N2 | 1.9420 (17) | C9—C8 | 1.390 (3) |
| Cu1—N1 | 2.0397 (18) | C15—H15 | 0.9500 |
| S1—O1 | 1.4466 (17) | C15—C14 | 1.394 (3) |
| S1—O3 | 1.4409 (18) | C14—H14 | 0.9500 |
| S1—O2 | 1.4392 (17) | C14—C13 | 1.376 (3) |
| S1—C16 | 1.826 (2) | C12—H12 | 0.9500 |
| F2—C16 | 1.331 (3) | C12—C13 | 1.401 (3) |
| F1—C16 | 1.335 (3) | C13—H13 | 0.9500 |
| F3—C16 | 1.337 (3) | C7—H7 | 0.9500 |
| N3—C11 | 1.362 (3) | C7—C8 | 1.392 (3) |
| N3—C15 | 1.339 (3) | C4—H4 | 0.9500 |
| N2—C10 | 1.335 (3) | C4—C3 | 1.391 (3) |
| N2—C6 | 1.336 (3) | C8—H8 | 0.9500 |
| N1—C5 | 1.364 (3) | C2—H2 | 0.9500 |
| N1—C1 | 1.336 (3) | C2—C1 | 1.384 (3) |
| C11—C10 | 1.481 (3) | C2—C3 | 1.386 (3) |
| C11—C12 | 1.380 (3) | C1—H1 | 0.9500 |
| C5—C6 | 1.479 (3) | C3—H3 | 0.9500 |
| C5—C4 | 1.388 (3) | ||
| Cl1—Cu1—Cl1i | 89.944 (18) | C8—C9—C10 | 117.9 (2) |
| N3—Cu1—Cl1i | 90.30 (5) | C8—C9—H9 | 121.0 |
| N3—Cu1—Cl1 | 99.82 (5) | N3—C15—H15 | 118.9 |
| N3—Cu1—N1 | 159.58 (7) | N3—C15—C14 | 122.2 (2) |
| N2—Cu1—Cl1i | 90.83 (5) | C14—C15—H15 | 118.9 |
| N2—Cu1—Cl1 | 179.20 (5) | C15—C14—H14 | 120.4 |
| N2—Cu1—N3 | 80.39 (7) | C13—C14—C15 | 119.1 (2) |
| N2—Cu1—N1 | 80.11 (7) | C13—C14—H14 | 120.4 |
| N1—Cu1—Cl1 | 99.60 (5) | C11—C12—H12 | 120.7 |
| N1—Cu1—Cl1i | 95.97 (5) | C11—C12—C13 | 118.7 (2) |
| Cu1—Cl1—Cu1i | 90.056 (18) | C13—C12—H12 | 120.7 |
| O1—S1—C16 | 102.05 (10) | C14—C13—C12 | 119.2 (2) |
| O3—S1—O1 | 114.97 (10) | C14—C13—H13 | 120.4 |
| O3—S1—C16 | 103.57 (11) | C12—C13—H13 | 120.4 |
| O2—S1—O1 | 114.19 (11) | C6—C7—H7 | 121.0 |
| O2—S1—O3 | 115.73 (12) | C6—C7—C8 | 118.1 (2) |
| O2—S1—C16 | 103.90 (11) | C8—C7—H7 | 121.0 |
| C11—N3—Cu1 | 113.61 (14) | C5—C4—H4 | 120.6 |
| C15—N3—Cu1 | 127.30 (15) | C5—C4—C3 | 118.8 (2) |
| C15—N3—C11 | 118.61 (19) | C3—C4—H4 | 120.6 |
| C10—N2—Cu1 | 118.38 (14) | C9—C8—C7 | 121.0 (2) |
| C10—N2—C6 | 123.04 (18) | C9—C8—H8 | 119.5 |
| C6—N2—Cu1 | 118.58 (14) | C7—C8—H8 | 119.5 |
| C5—N1—Cu1 | 113.57 (14) | C1—C2—H2 | 120.5 |
| C1—N1—Cu1 | 127.51 (15) | C1—C2—C3 | 119.1 (2) |
| C1—N1—C5 | 118.60 (19) | C3—C2—H2 | 120.5 |
| N3—C11—C10 | 114.06 (19) | F2—C16—S1 | 111.78 (16) |
| N3—C11—C12 | 122.2 (2) | F2—C16—F1 | 107.3 (2) |
| C12—C11—C10 | 123.8 (2) | F2—C16—F3 | 107.8 (2) |
| N1—C5—C6 | 114.01 (18) | F1—C16—S1 | 110.99 (16) |
| N1—C5—C4 | 121.9 (2) | F1—C16—F3 | 106.5 (2) |
| C4—C5—C6 | 124.13 (19) | F3—C16—S1 | 112.25 (17) |
| N2—C10—C11 | 113.09 (18) | N1—C1—C2 | 122.5 (2) |
| N2—C10—C9 | 119.91 (19) | N1—C1—H1 | 118.7 |
| C9—C10—C11 | 127.0 (2) | C2—C1—H1 | 118.7 |
| N2—C6—C5 | 113.29 (18) | C4—C3—H3 | 120.5 |
| N2—C6—C7 | 120.03 (19) | C2—C3—C4 | 119.1 (2) |
| C7—C6—C5 | 126.68 (19) | C2—C3—H3 | 120.5 |
| C10—C9—H9 | 121.0 | ||
| Cu1—N3—C11—C10 | −7.7 (2) | N1—C5—C4—C3 | 0.2 (3) |
| Cu1—N3—C11—C12 | 170.55 (16) | C11—N3—C15—C14 | 1.5 (3) |
| Cu1—N3—C15—C14 | −170.07 (15) | C11—C10—C9—C8 | −178.6 (2) |
| Cu1—N2—C10—C11 | −0.7 (2) | C11—C12—C13—C14 | 0.8 (3) |
| Cu1—N2—C10—C9 | 179.33 (15) | C5—N1—C1—C2 | −1.1 (3) |
| Cu1—N2—C6—C5 | −1.4 (2) | C5—C6—C7—C8 | −177.9 (2) |
| Cu1—N2—C6—C7 | 179.33 (15) | C5—C4—C3—C2 | −1.7 (3) |
| Cu1—N1—C5—C6 | 7.0 (2) | C10—N2—C6—C5 | 179.27 (18) |
| Cu1—N1—C5—C4 | −172.78 (16) | C10—N2—C6—C7 | 0.0 (3) |
| Cu1—N1—C1—C2 | 171.97 (16) | C10—C11—C12—C13 | 178.98 (19) |
| O1—S1—C16—F2 | −60.24 (18) | C10—C9—C8—C7 | −0.1 (3) |
| O1—S1—C16—F1 | 59.49 (19) | C6—N2—C10—C11 | 178.63 (18) |
| O1—S1—C16—F3 | 178.50 (17) | C6—N2—C10—C9 | −1.4 (3) |
| O3—S1—C16—F2 | −179.95 (16) | C6—C5—C4—C3 | −179.6 (2) |
| O3—S1—C16—F1 | −60.22 (19) | C6—C7—C8—C9 | −1.1 (3) |
| O3—S1—C16—F3 | 58.8 (2) | C15—N3—C11—C10 | 179.69 (18) |
| O2—S1—C16—F2 | 58.73 (19) | C15—N3—C11—C12 | −2.1 (3) |
| O2—S1—C16—F1 | 178.47 (17) | C15—C14—C13—C12 | −1.5 (3) |
| O2—S1—C16—F3 | −62.5 (2) | C12—C11—C10—N2 | −172.60 (19) |
| N3—C11—C10—N2 | 5.6 (3) | C12—C11—C10—C9 | 7.4 (3) |
| N3—C11—C10—C9 | −174.4 (2) | C4—C5—C6—N2 | 175.9 (2) |
| N3—C11—C12—C13 | 1.0 (3) | C4—C5—C6—C7 | −4.9 (3) |
| N3—C15—C14—C13 | 0.3 (3) | C1—N1—C5—C6 | −178.99 (18) |
| N2—C10—C9—C8 | 1.4 (3) | C1—N1—C5—C4 | 1.3 (3) |
| N2—C6—C7—C8 | 1.2 (3) | C1—C2—C3—C4 | 1.9 (3) |
| N1—C5—C6—N2 | −3.8 (3) | C3—C2—C1—N1 | −0.4 (3) |
| N1—C5—C6—C7 | 175.3 (2) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Acknowledgements
We are thankful for the support of the Department of Chemistry and Biochemistry at the University of the Incarnate Word and the X-ray Diffraction Laboratory at The University of Texas at San Antonio.
Funding information
Funding for this research was provided by: Welch Foundation (award No. BN0032).
References
 Choroba, K., Machura, B., Kula, S., Raposo, L. R., Fernandes, A. R., Kruszynski, R., Erfurt, K., Shul'pina, L. S., Kozlov, Y. N. & Shul'pin, G. B. (2019). Dalton Trans. 48, 12656–12673. Web of Science CSD CrossRef CAS PubMed Google Scholar
Choroba, K., Machura, B., Kula, S., Raposo, L. R., Fernandes, A. R., Kruszynski, R., Erfurt, K., Shul'pina, L. S., Kozlov, Y. N. & Shul'pin, G. B. (2019). Dalton Trans. 48, 12656–12673. Web of Science CSD CrossRef CAS PubMed Google Scholar
 Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341. Web of Science CrossRef CAS IUCr Journals Google Scholar
Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341. Web of Science CrossRef CAS IUCr Journals Google Scholar
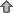 Gasser, G., Labat, G. & Stoeckli-Evans, H. (2004). Acta Cryst. E60, m244–m246. CSD CrossRef IUCr Journals Google Scholar
Gasser, G., Labat, G. & Stoeckli-Evans, H. (2004). Acta Cryst. E60, m244–m246. CSD CrossRef IUCr Journals Google Scholar
 Glišić, B. Đ., Nikodinovic-Runic, J., Ilic-Tomic, T., Wadepohl, H., Veselinović, A., Opsenica, I. M. & Djuran, M. I. (2018). Polyhedron, 139, 313–322. Google Scholar
Glišić, B. Đ., Nikodinovic-Runic, J., Ilic-Tomic, T., Wadepohl, H., Veselinović, A., Opsenica, I. M. & Djuran, M. I. (2018). Polyhedron, 139, 313–322. Google Scholar
 Karges, J., Xiong, K., Blacque, O., Chao, H. & Gasser, G. (2021). Inorg. Chim. Acta, 516, 120137. CSD CrossRef Google Scholar
Karges, J., Xiong, K., Blacque, O., Chao, H. & Gasser, G. (2021). Inorg. Chim. Acta, 516, 120137. CSD CrossRef Google Scholar
 Li, C., Xu, F., Zhao, Y., Zheng, W., Zeng, W., Luo, Q., Wang, Z., Wu, K., Du, J. & Wang, F. (2020). Front. Chem. 8, 210. CrossRef PubMed Google Scholar
Li, C., Xu, F., Zhao, Y., Zheng, W., Zeng, W., Luo, Q., Wang, Z., Wu, K., Du, J. & Wang, F. (2020). Front. Chem. 8, 210. CrossRef PubMed Google Scholar
 Malarz, K., Zych, D., Gawecki, R., Kuczak, M., Musioł, R. & Mrozek-Wilczkiewicz, A. (2021). Eur. J. Med. Chem. 212, 113032. CrossRef PubMed Google Scholar
Malarz, K., Zych, D., Gawecki, R., Kuczak, M., Musioł, R. & Mrozek-Wilczkiewicz, A. (2021). Eur. J. Med. Chem. 212, 113032. CrossRef PubMed Google Scholar
 Rigaku OD (2019). CrysAlis PRO. Rigaku Oxford Diffraction, Rigaku Corporation, Oxford, England. Google Scholar
Rigaku OD (2019). CrysAlis PRO. Rigaku Oxford Diffraction, Rigaku Corporation, Oxford, England. Google Scholar
 Rojo, T., Arriortua, M. I., Ruiz, J., Darriet, J., Villeneuve, G. & Beltran-Porter, D. (1987). J. Chem. Soc. Dalton Trans. pp. 285–291. CSD CrossRef Web of Science Google Scholar
Rojo, T., Arriortua, M. I., Ruiz, J., Darriet, J., Villeneuve, G. & Beltran-Porter, D. (1987). J. Chem. Soc. Dalton Trans. pp. 285–291. CSD CrossRef Web of Science Google Scholar
 Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8. Web of Science CrossRef IUCr Journals Google Scholar
Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8. Web of Science CrossRef IUCr Journals Google Scholar
 Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8. Web of Science CrossRef IUCr Journals Google Scholar
Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8. Web of Science CrossRef IUCr Journals Google Scholar
 Valdés-Martínez, J., Salazar-Mendoza, D. & Toscano, R. A. (2002). Acta Cryst. E58, m712–m714. Web of Science CSD CrossRef IUCr Journals Google Scholar
Valdés-Martínez, J., Salazar-Mendoza, D. & Toscano, R. A. (2002). Acta Cryst. E58, m712–m714. Web of Science CSD CrossRef IUCr Journals Google Scholar
 Wei, C., He, Y., Shi, X. & Song, Z. (2019). Coord. Chem. Rev. 385, 1–19. CrossRef CAS PubMed Google Scholar
Wei, C., He, Y., Shi, X. & Song, Z. (2019). Coord. Chem. Rev. 385, 1–19. CrossRef CAS PubMed Google Scholar
This is an open-access article distributed under the terms of the Creative Commons Attribution (CC-BY) Licence, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are cited.


 journal menu
journal menu











 access
access


