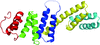issue contents
February 2012 issue
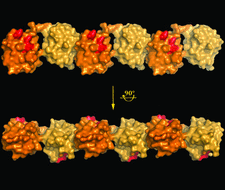
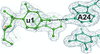
Cover illustration: A surface representation of two adjacent linear diubiquitin molecules in two views related by a 90° rotation around the horizontal axis (p. 102). The distal ubiquitins are denoted in darker orange, whereas the proximal moieties are shown in tan. The hydrophobic patches (Leu8, Ile44 and Val70) are shown in red and are aligned on the same side in every other moiety along the straight line of linear diubiquitin chains in the crystal.
research papers
BamD is part of the Escherichia coli outer membrane protein complex (BAM complex) and is essential for the survival of E. coli. The structure of BamD at 2.6 Å resolution shows that this lipoprotein is composed of ten α-helices that form five tetratricopeptide-repeat (TPR) motifs.
PDB reference: BamD, 3q5m
A new crystal structure of linear diubiquitin adopts a compact conformation that differs from its previously reported extended conformation.
PDB reference: diubiquitin, 3axc
Download citation


Download citation


The first structure of a subfamily 4 peptaibol antibiotic, trichovirin I-4A, is described at atomic resolution. The structure suggests a conformational transition to obtain the necessary amphiphilicity for membrane insertion and water/ion transport across the hydrophobic barrier.
PDB reference: trichovirin I-4A, 3sbn
The 1.75 Å resolution X-ray crystallographic structure of human evectin-2 pleckstrin homology domain revealed ligand-induced conformational change. This structural change effectively explains the strict phospholipid binding specificity.
PDB reference: human evectin-2 PH domain, 3via
Approximately half of global radiation damage to thaumatin crystals can be outrun at 260 K if data are collected in less than 1 s.
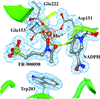
The structure of M. tuberculosis DXR (also known as IspC) in complex with the antibiotic FR-900098, NADPH and manganese was determined. This new crystal form diffracts to the highest resolution (1.65 Å) reported to date for this enzyme from any species.
PDB reference: MtDXR–Mn–F98–NADPH, 4a03
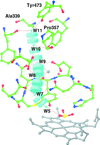
An unusual covalent bond between the side chains of the catalytic Tyr residue and Gln decreases the activity of cytochrome c nitrite reductase. The Tyr residue is a proton donor during the catalysis.
PDB references: TvNiR–NO2, 3rkh; TvNiRb–PO4, 3sce; TvNiRb–SO3, 3lgq; TvNiRb–SO3(MV), 3lg1; TvNiRb–NH2OH, 3s7w; TvNiRb–H2O, 3uu9
Open  access
access
 access
accessThe human Kif7 motor domain structure provides insights into a kinesin of medical significance.
Crystal structures of the bacterial α1,6-fucosyltransferase NodZ in complex with GDP and GDP-fucose are presented.
The high-resolution crystal structure of an RNA/DNA dodecamer with the HIV-1 polypurine-tract sequence reveals an A-type hybrid duplex with a complete absence of any C2′-endo ribonucleotides and with segments of increased flexibility along the RNA chain.
The crystal structure of the effector-binding domain of the transcriptional repressor AraR from B. subtilis in complex with the effector molecule (L-arabinose) was determined at 2.2 Å resolution. A detailed analysis of the crystal identified a dimer organization that is distinctive from that of other members of the GalR/LacI family.
PDB reference: AraR effector-binding domain, 3tb6
The role of Asp116 in the pronotation events taking place during CotA laccase catalytic mechanism was investigated. The crystal structure determination of three distinct mutants (D116A, D116N and D116E), produced by site-saturation mutagenesis, together with theoretical calculations have provided evidence of its importance during the reductive cleavage of dioxygen to water.
international union of crystallography
Free 



 journal menu
journal menu