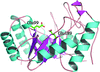issue contents
August 2015 issue
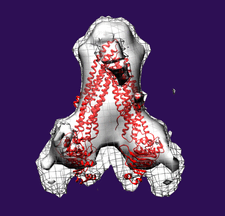
Cover illustration: Subtomogram average side-view of the human breast cancer resistance protein ABCG2 homodimer. Two ABCG2 monomers form a V-shaped dimer with an angle of approximately 60° between their long axes (Rosenberg et al., p. 1725).
research papers
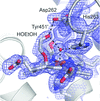
Transthyretin represents a notable example of an amyloidogenic protein which undergoes misfolding, leading to a degenerative disease known as amyloidosis. Evidence is provided for significant conformational differences between the two thyroxine binding sites in solution, consistent with an asymmetric ligand-binding pattern. Rationalization of the structural basis of the observed asymmetry will aid in the design of new drugs effective as transthyretin stabilizers, with the prospect of their use in the pharmacological therapy of transthyretin amyloidosis.
Deerpox virus employs the cell-death inhibitor DPV022 to prevent premature host cell death by engaging pro-death Bcl-2 proteins via two independent ligand-binding sites as part of a domain-swapped Bcl-2 fold.
Open  access
access
 access
accessA method is presented to detect peptide bonds that need either a trans–cis flip or a peptide-plane flip.
The LpfD protein represents the tip adhesin of enterobacteriaceal long polar fimbriae (LPF), and LpfD from the adherent invasive E. coli strain LF82 directs LPF binding towards the intestinal epithelium and underlying tissue.
PDB reference: LpfD, 5afo
Open  access
access
 access
accessAn alternative strategy for PEG sampling is suggested through the use of four newly defined PEG smears to enhance chemical space in reduced screens with a benefit towards protein crystallization.
The crystal structures of substrate and nucleotide bound propionate kinase and its site-directed mutants at a resolution of 1.8–2.0 Å are presented here. A probable catalytic mechanism is proposed which may be applicable to acetokinase family members.
Crystal structures of the eukaryotic DNA replication initiator Sld7 in complex with Sld3 from yeasts were determined. The quaternary structure of the Sld3–Sld7 complex appears to be suitable to activate two helicase molecules loaded onto replication origins in a head-to-head manner.
Open  access
access
 access
accessA method of simulating X-ray diffuse scattering from multi-model PDB files is presented. Despite similar agreement with Bragg data, different translation–libration–screw refinement strategies produce unique diffuse intensity patterns.
Open  access
access
 access
accessProcedures are described for extracting the vibration and libration parameters corresponding to a given set of TLS matrices and their simultaneous validation. Knowledge of these parameters allows the generation of structural ensembles corresponding to these matrices.
A crystallographic cocktail screening campaign against 680 fragments revealed a binding hotspot in the structure of T. cruzi histidyl-tRNA synthetase.
PDB references: Trypanosoma cruzi HisRS, complex with Chem 79, 4yp0; complex with Chem 84, 4ypf; complex with Chem 89, 4yrc; complex with Chem 145, 4yre; complex with Chem 148, 4yrf; complex with Chem 149, 4yrg; complex with Chem 166, 4yri; complex with Chem 256, 4yrj; complex with Chem 262, 4yrl; complex with Chem 260, 4yrk; complex with Chem 443, 4yrm; complex with Chem 475, 4yrn; complex with Chem 491, 4yro; complex with Chem 707, 4yrp; complex with Chem 744, 4yrq; complex with Chem 1691, 4yrr; complex with Chem 1698, 4yrs; complex with Chem 1781, 4yrt
The three-dimensional structure of the green fluorescent protein NowGFP (a successor to Cerulean) with an anionic tryptophan-based chromophore and that of the photoconverted form NowGFP_conv are reported. The relationship between structure and photophysical characteristics is investigated.
An unexpected reactivity is found in a unique haem moiety bound covalently to an oxidase.
PDB reference: hydroxylamine oxidoreductase, 4rwm
The crystal structures of a novel glycoside hydrolase (GH) family 17 β-1,3-glucanosyltransferase from R. miehei (RmBgt17A) and two different substrate complexes have been solved at resolutions of 1.30, 2.30 and 2.27 Å, respectively. The first crystal structure of a GH family 17 β-1,3-glucanosyltransferase may be useful in studies of the catalytic mechanism of GH family 17 proteins and provides a basis for further enzymatic engineering or antifungal drug screening.
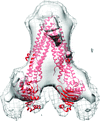
The first three-dimensional structure of human ABCG2 in the absence of nucleotides and transported substrates has been determined at 2.0 nm resolution. In this state, ABCG2 is in an inward-facing conformation.
Shikimate kinase was determined to be essential for the growth and survival of A. baumannii in a rat soft-tissue infection model. The crystal structure of this enzyme in complex with shikimate and sulfate ion was determined to 1.91 Å resolution, revealing ligand-induced conformational changes of key motifs.
The crystal structure of the α-carbonic anhydrase (α-CA) from the gammaproteobacterium T. crunogena XCL-2 has been determined to 2.6 Å resolution and reveals features that provide it with thermal and pH stability that are not seen in human α-CAs.
PDB reference: α-carbonic anhydrase, 4xz5
Open  access
access
 access
accessA fully automatic system has been developed that performs X-ray centring and characterization of, and data collection from, large numbers of cryocooled crystals without human intervention.
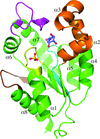
SaeR can bind to P1 promoter DNA using the canonical winged helix–turn–helix module.
PDB reference: DNA-binding domain of SaeR, 4qwq
In order to accelerate ligand screening using X-ray crystallography, the combination of in situ diffraction with pre-coating of 96-well crystallization plates using a DMSO stock solution of putative ligands before performing protein crystallization is proposed.
PDB references: human nuclear receptor PPARγ, complex with rosiglitazone, 4xld; hen egg-white lysozyme, complex with benzamidine, 4xn6; human cyclophilin D, complex with ethyl 2-({[(4-aminophenyl)methyl]carbamoyl}amino)acetate, room temperature, 4xnc; 4zsc; 100 K, 3rdc; complex with 1-(4-aminobenzyl)-3-[4-(methylthio)-1-{2-[2-(methylthio)]phenyl}pyrrolidin-1-yl)-1-oxobutan-2-yl]urea, 100 K, 4j5c; room temperature, 4zsd; Erk-2, complex with 1-phenyl-1H-1,2,4-triazole-3,5-diamine, room temperature, 4xne; 100 K, 4xp2; complex with 2-amino-6-thiopurine, room temperature, 4xoy; 100 K, 4xp3; complex with 1-N-{[3-(benzyloxy)phenyl]methyl}-2H-1,2,3,4-tetrazole-1,5-diamine, room temperature, 4xrj; 100 K, 4xoz; complex with 3-cyano-7-azaindole, room temperature, 4xrl; 100 K, 4xp0


 journal menu
journal menu