issue contents
February 2020 issue
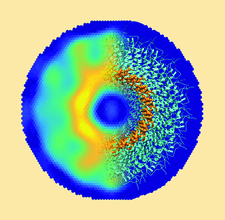
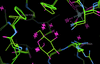
Cover illustration: FiXR: a framework to reconstruct fiber cross sections from X-ray fiber diffraction experiments [Roig-Solvas et al. (2020), Acta Cryst. D76, 102-117]. A FiXR reconstruction of tobacco mosaic virus using SAXS measurements is compared side-by-side with PDB entry 2tmv.
ISDSB2019
Open  access
access
 access
accessThe three probes of the structure of matter (X-rays, neutrons and electrons) in biology have complementary properties and strengths. For the study of combined states of order and disorder, NMR crystallography is also applicable. Overall, to understand a biological system, the requirements are physiologically relevant results. Reporting of protein crystal structures, and their associated database entries, could usefully indicate how close to the biological situation they are.
CCP-EM
Open  access
access
 access
accessAmyloid structure determination in RELION is increasingly popular, but may suffer from refinements becoming stuck in local minima. This paper describes a new method for initial model generation that may alleviate this problem.
research papers
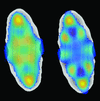
A method is presented to reconstruct the 2D electron densities of fiber cross-sections from disordered small-angle X-ray scattering measurements, combining fiber diffraction theory with physical constraints, like finite cross-section size and non-negative density, in a numerical optimization framework.
Neutron and X-ray diffraction were used to investigate the effects of a Glu166Gln mutation on the active site of a class A β-lactamase.
PDB reference: Toho-1, Glu166Gln mutant, 6u58
The first structure of a thrombospondin module 1 (TSP1) domain from a CCN-family matricellular protein has been determined by X-ray crystallography. The structure shows a typical three-stranded fold, but lacks the π-stacked structure that is usually found in these domains. The structure reveals highest conservation in the positively charged central segment, which is predicted to be a binding site for heparan sulfates. The atypical features of this domain have been used to revise the definition of TSP1 domains and to identify a number of new domains in sequence databases.
PDB reference: thrombospondin module 1 domain, 6rk1
Three non-aminoacyl-tRNA synthetase (aaRS) scaffold proteins, AIMP1/p43 (pro-EMAP II), AIMP2/p38 and AIMP3/p18, known as aaRS-interacting multifunctional proteins, are critical in driving the assembly of multi-aaRS complexes in higher eukaryotes. Here, the structural features of the N- and C-terminal domains of the Plasmodium vivax p43 (Pvp43) protein are addressed. A comprehensive structural analyses of the two domains of Pvp43 reveals several notable features which suggest that it is likely to be a soluble, cytosolic protein with a dimeric assembly of its N-terminal GST-like domain that differs from canonical GST dimers.
Open  access
access
 access
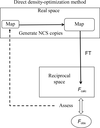
accessA noncrystallographic symmetry-constrained map obtained by direct density optimization is efficient and equivalent to a noncrystallographic symmetry-averaging map. Using the noncrystallographic symmetry-constrained map, the structure of a T = 1 Penaeus vannamei nodavirus shell-domain subviral particle was newly determined by including twinned data.
Open  access
access
 access
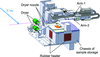
accessA rapid and reliable sample changer, SPACE-II, has been developed at the SPring-8 macromolecular crystallography beamline BL41XU. It enables samples to be exchanged in 16 s, of which its action accounts for only 11 s. Two years of operating SPACE-II demonstrated that the average number of sample exchanges per day was increased by 40% compared with the previous model, and it had an error rate of only 0.089%.
Open  access
access
 access
accessThe structure of Streptococcus pneumoniae TrpA, which is the α subunit of tryptophan synthase, reveals 3D domain swapping, which has never before been observed for a TIM-barrel protein.
PDB reference: domain-swapped TrpA, 6qky
Open  access
access
 access
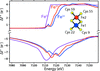
accessElectronic configurations at distinct metal centers within a metalloprotein may be characterized by inspecting the scattering factors at the X-ray absorption edge. Such experiments may be feasible at XFEL sources, using Bayesian data analysis.


 journal menu
journal menu