In the title compound, [Cu(C
8H
4O
4)(C
14H
8N
4)(H
2O)]·C
3H
7NO·H
2O, the Cu
II atom binds a water molecule and is chelated by the pyrazinophenanthroline
N-heterocycle. It is also linked covalently to a terephthalate dianion that lies on a special position of
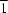
site symmetry. This dianion serves as a spacer to connect adjacent square-pyramidal metal atoms into a helical chain that propagates along the
b axis of the monoclinic unit cell. The chain motif is consolidated into a layer structure by hydrogen bonds involving the coordinated and uncoordinated water molecules. The Cu atom and the coordinated water molecule lie on a mirror plane, while the
N-heterocycle, the disordered uncoordinated water molecule and disordered dimethylformamide solvent molecule lie about a mirror plane.
Supporting information
CCDC reference: 630466
Key indicators
- Single-crystal X-ray study
- T = 293 K
- Mean
![[sigma]](https://journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.005 Å
(C-C) = 0.005 Å
- Disorder in main residue
- R factor = 0.050
- wR factor = 0.144
- Data-to-parameter ratio = 13.3
checkCIF/PLATON results
No syntax errors found
 Alert level A
DIFF020_ALERT_1_A _diffrn_standards_interval_count and
_diffrn_standards_interval_time are missing. Number of measurements
between standards or time (min) between standards.
DIFF022_ALERT_1_A _diffrn_standards_decay_% is missing
Percentage decrease in standards intensity.
PLAT780_ALERT_1_A Coordinates do not Form a Properly Connected Set ?
Alert level A
DIFF020_ALERT_1_A _diffrn_standards_interval_count and
_diffrn_standards_interval_time are missing. Number of measurements
between standards or time (min) between standards.
DIFF022_ALERT_1_A _diffrn_standards_decay_% is missing
Percentage decrease in standards intensity.
PLAT780_ALERT_1_A Coordinates do not Form a Properly Connected Set ?
 Alert level B
PLAT415_ALERT_2_B Short Inter D-H..H-X H1W1 .. H13C .. 2.03 Ang.
PLAT415_ALERT_2_B Short Inter D-H..H-X H1W1 .. H13C .. 2.03 Ang.
PLAT416_ALERT_2_B Short Intra D-H..H-D H1W2 .. H2W2 .. 1.86 Ang.
PLAT416_ALERT_2_B Short Intra D-H..H-D H1W2 .. H2W2 .. 1.86 Ang.
Alert level B
PLAT415_ALERT_2_B Short Inter D-H..H-X H1W1 .. H13C .. 2.03 Ang.
PLAT415_ALERT_2_B Short Inter D-H..H-X H1W1 .. H13C .. 2.03 Ang.
PLAT416_ALERT_2_B Short Intra D-H..H-D H1W2 .. H2W2 .. 1.86 Ang.
PLAT416_ALERT_2_B Short Intra D-H..H-D H1W2 .. H2W2 .. 1.86 Ang.
 Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.701 0.909
Tmin(prime) and Tmax expected: 0.811 0.908
RR(prime) = 0.863
Please check that your absorption correction is appropriate.
PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ?
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 0.50 Ratio
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.86
PLAT150_ALERT_1_C Volume as Calculated Differs from that Given ... 1197.00 Ang-3
PLAT152_ALERT_1_C Supplied and Calc Volume s.u. Inconsistent ..... ?
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for N2
PLAT301_ALERT_3_C Main Residue Disorder ......................... 5.00 Perc.
PLAT720_ALERT_4_C Number of Unusual/Non-Standard Label(s) ........ 4
PLAT731_ALERT_1_C Bond Calc 0.84(4), Rep 0.850(10) ...... 4.00 su-Ra
O1W -H1W2 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.84(4), Rep 0.850(10) ...... 4.00 su-Ra
O1W -H2# 1.555 1.555
Alert level C
ABSTM02_ALERT_3_C The ratio of expected to reported Tmax/Tmin(RR') is < 0.90
Tmin and Tmax reported: 0.701 0.909
Tmin(prime) and Tmax expected: 0.811 0.908
RR(prime) = 0.863
Please check that your absorption correction is appropriate.
PLAT041_ALERT_1_C Calc. and Rep. SumFormula Strings Differ .... ?
PLAT042_ALERT_1_C Calc. and Rep. MoietyFormula Strings Differ .... ?
PLAT045_ALERT_1_C Calculated and Reported Z Differ by ............ 0.50 Ratio
PLAT061_ALERT_3_C Tmax/Tmin Range Test RR' too Large ............. 0.86
PLAT150_ALERT_1_C Volume as Calculated Differs from that Given ... 1197.00 Ang-3
PLAT152_ALERT_1_C Supplied and Calc Volume s.u. Inconsistent ..... ?
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for O1
PLAT241_ALERT_2_C Check High Ueq as Compared to Neighbors for N2
PLAT301_ALERT_3_C Main Residue Disorder ......................... 5.00 Perc.
PLAT720_ALERT_4_C Number of Unusual/Non-Standard Label(s) ........ 4
PLAT731_ALERT_1_C Bond Calc 0.84(4), Rep 0.850(10) ...... 4.00 su-Ra
O1W -H1W2 1.555 1.555
PLAT735_ALERT_1_C D-H Calc 0.84(4), Rep 0.850(10) ...... 4.00 su-Ra
O1W -H2# 1.555 1.555
 Alert level G
PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K
PLAT200_ALERT_1_G Check the Reported _diffrn_ambient_temperature . 293 K
Alert level G
PLAT199_ALERT_1_G Check the Reported _cell_measurement_temperature 293 K
PLAT200_ALERT_1_G Check the Reported _diffrn_ambient_temperature . 293 K
3 ALERT level A = In general: serious problem
4 ALERT level B = Potentially serious problem
13 ALERT level C = Check and explain
2 ALERT level G = General alerts; check
12 ALERT type 1 CIF construction/syntax error, inconsistent or missing data
6 ALERT type 2 Indicator that the structure model may be wrong or deficient
3 ALERT type 3 Indicator that the structure quality may be low
1 ALERT type 4 Improvement, methodology, query or suggestion
0 ALERT type 5 Informative message, check
Data collection: PROCESS-AUTO (Rigaku, 1998); cell refinement: PROCESS-AUTO; data reduction: PROCESS-AUTO; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: X-SEED (Barbour, 2001); software used to prepare material for publication: publCIF (Westrip, 2006).
catena-Poly[[[aqua(pyrazino[2,3-
f][1,10]phenanthroline-
κ2N,
N')copper(II)]-µ-terephthalato-
κ2O:
O']
N,
N-dimethylformamide monohydrate]
top
Crystal data top
| [Cu(C8H4O4)(C14H8N4)(H2O)]·C3H7NO·H2O | F(000) = 586 |
| Mr = 569.02 | Dx = 1.578 Mg m−3 |
| Monoclinic, P21/m | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yb | Cell parameters from 9444 reflections |
| a = 7.275 (4) Å | θ = 3.2–27.5° |
| b = 13.970 (5) Å | µ = 0.97 mm−1 |
| c = 11.794 (5) Å | T = 293 K |
| β = 92.78 (2)° | Block, green |
| V = 1197 (1) Å3 | 0.21 × 0.19 × 0.10 mm |
| Z = 2 | |
Data collection top
Rigaku R-AXIS RAPID IP
diffractometer | 2856 independent reflections |
| Radiation source: fine-focus sealed tube | 2229 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.034 |
| ω scans | θmax = 27.5°, θmin = 3.2° |
Absorption correction: multi-scan
(ABSCOR; Higashi, 1995) | h = −9→9 |
| Tmin = 0.701, Tmax = 0.909 | k = −16→18 |
| 11896 measured reflections | l = −15→14 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.144 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.077P)2 + 0.8938P]
where P = (Fo2 + 2Fc2)/3 |
| 2856 reflections | (Δ/σ)max = 0.001 |
| 214 parameters | Δρmax = 0.85 e Å−3 |
| 58 restraints | Δρmin = −0.56 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Cu1 | 0.35210 (8) | 0.2500 | 0.64318 (4) | 0.0386 (2) | |
| O1 | 0.4201 (4) | 0.3484 (2) | 0.7528 (2) | 0.0575 (7) | |
| O2 | 0.1446 (4) | 0.3936 (2) | 0.8086 (2) | 0.0617 (8) | |
| O1W | 0.6390 (5) | 0.2500 | 0.5738 (3) | 0.0607 (11) | |
| H1W1 | 0.662 (7) | 0.2500 | 0.5043 (13) | 0.073* | |
| H1W2 | 0.738 (4) | 0.2500 | 0.614 (4) | 0.073* | |
| O2W | 0.9645 (8) | 0.260 (4) | 0.6961 (5) | 0.070 (7) | 0.50 |
| H2W1 | 1.036 (10) | 0.302 (6) | 0.726 (7) | 0.084* | 0.50 |
| H2W2 | 0.889 (10) | 0.242 (9) | 0.745 (5) | 0.084* | 0.50 |
| N1 | 0.2637 (4) | 0.34436 (18) | 0.5218 (2) | 0.0375 (6) | |
| N2 | 0.0266 (5) | 0.3496 (3) | 0.1299 (3) | 0.0635 (10) | |
| C1 | 0.3144 (5) | 0.3923 (2) | 0.8172 (3) | 0.0433 (8) | |
| C2 | 0.4119 (5) | 0.4492 (2) | 0.9116 (2) | 0.0371 (7) | |
| C3 | 0.6013 (5) | 0.4521 (2) | 0.9218 (3) | 0.0429 (8) | |
| H3 | 0.6701 | 0.4200 | 0.8693 | 0.051* | |
| C4 | 0.6901 (5) | 0.5024 (2) | 1.0098 (3) | 0.0419 (7) | |
| H4 | 0.8180 | 0.5039 | 1.0161 | 0.050* | |
| C5 | 0.1969 (4) | 0.30106 (19) | 0.4258 (2) | 0.0328 (6) | |
| C6 | 0.1358 (4) | 0.3528 (2) | 0.3294 (3) | 0.0406 (7) | |
| C7 | 0.1431 (6) | 0.4526 (3) | 0.3350 (3) | 0.0565 (10) | |
| H7 | 0.1010 | 0.4894 | 0.2734 | 0.068* | |
| C8 | 0.2127 (7) | 0.4954 (3) | 0.4321 (3) | 0.0641 (12) | |
| H8 | 0.2206 | 0.5618 | 0.4365 | 0.077* | |
| C9 | 0.2720 (6) | 0.4390 (2) | 0.5243 (3) | 0.0523 (9) | |
| H9 | 0.3189 | 0.4690 | 0.5899 | 0.063* | |
| C10 | 0.0781 (5) | 0.2997 (2) | 0.2291 (2) | 0.0434 (8) | |
| C11 | −0.0069 (5) | 0.2992 (2) | 0.0540 (3) | 0.0437 (8) | |
| H11 | −0.0374 | 0.3286 | −0.0151 | 0.052* | |
| O5 | 0.6708 (10) | 0.1582 (4) | 0.3734 (5) | 0.0651 (17) | 0.50 |
| N3 | 0.5780 (5) | 0.2533 (16) | 0.2253 (4) | 0.0453 (12) | 0.50 |
| C12 | 0.6065 (15) | 0.1672 (18) | 0.2728 (10) | 0.050 (4) | 0.50 |
| H12 | 0.5782 | 0.1126 | 0.2304 | 0.060* | 0.50 |
| C13 | 0.622 (2) | 0.3392 (17) | 0.2888 (12) | 0.054 (4) | 0.50 |
| H13A | 0.5098 | 0.3705 | 0.3079 | 0.080* | 0.50 |
| H13B | 0.6925 | 0.3813 | 0.2438 | 0.080* | 0.50 |
| H13C | 0.6917 | 0.3229 | 0.3572 | 0.080* | 0.50 |
| C14 | 0.5053 (9) | 0.2628 (15) | 0.1094 (5) | 0.055 (3) | 0.50 |
| H14A | 0.5976 | 0.2901 | 0.0637 | 0.082* | 0.50 |
| H14B | 0.3992 | 0.3038 | 0.1075 | 0.082* | 0.50 |
| H14C | 0.4709 | 0.2009 | 0.0802 | 0.082* | 0.50 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cu1 | 0.0572 (4) | 0.0337 (3) | 0.0237 (3) | 0.000 | −0.0115 (2) | 0.000 |
| O1 | 0.0702 (18) | 0.0646 (17) | 0.0363 (12) | 0.0002 (14) | −0.0127 (12) | −0.0222 (12) |
| O2 | 0.0586 (19) | 0.0634 (18) | 0.0611 (16) | −0.0045 (13) | −0.0199 (13) | −0.0174 (14) |
| O1W | 0.043 (2) | 0.096 (3) | 0.042 (2) | 0.000 | −0.0104 (16) | 0.000 |
| O2W | 0.071 (3) | 0.04 (2) | 0.094 (4) | 0.009 (7) | −0.046 (3) | 0.000 (5) |
| N1 | 0.0464 (16) | 0.0332 (13) | 0.0321 (13) | 0.0022 (11) | −0.0057 (11) | −0.0004 (10) |
| N2 | 0.059 (2) | 0.070 (2) | 0.062 (2) | 0.0172 (17) | 0.0069 (17) | 0.0347 (19) |
| C1 | 0.063 (2) | 0.0354 (16) | 0.0301 (15) | −0.0047 (15) | −0.0139 (15) | 0.0002 (12) |
| C2 | 0.0499 (19) | 0.0323 (15) | 0.0279 (14) | −0.0018 (13) | −0.0101 (13) | 0.0006 (11) |
| C3 | 0.055 (2) | 0.0411 (18) | 0.0320 (15) | 0.0001 (15) | −0.0023 (14) | −0.0053 (13) |
| C4 | 0.0433 (19) | 0.0434 (18) | 0.0384 (16) | −0.0021 (14) | −0.0059 (14) | −0.0036 (13) |
| C5 | 0.0315 (15) | 0.0406 (16) | 0.0258 (13) | 0.0017 (12) | −0.0026 (11) | 0.0037 (12) |
| C6 | 0.0361 (17) | 0.0518 (19) | 0.0338 (15) | 0.0054 (14) | −0.0001 (13) | 0.0084 (14) |
| C7 | 0.071 (3) | 0.048 (2) | 0.051 (2) | 0.0125 (18) | −0.0009 (19) | 0.0167 (17) |
| C8 | 0.100 (4) | 0.0336 (18) | 0.059 (2) | 0.006 (2) | 0.002 (2) | 0.0102 (17) |
| C9 | 0.077 (3) | 0.0325 (17) | 0.0471 (19) | −0.0009 (17) | −0.0033 (18) | −0.0013 (14) |
| C10 | 0.0347 (17) | 0.070 (2) | 0.0246 (14) | 0.0052 (15) | −0.0043 (12) | 0.0070 (14) |
| C11 | 0.0313 (17) | 0.0469 (19) | 0.053 (2) | 0.0046 (14) | 0.0034 (15) | 0.0080 (16) |
| O5 | 0.095 (5) | 0.050 (3) | 0.049 (3) | 0.014 (3) | 0.002 (3) | 0.003 (2) |
| N3 | 0.048 (2) | 0.043 (3) | 0.045 (2) | 0.003 (8) | 0.0017 (18) | −0.003 (8) |
| C12 | 0.056 (7) | 0.058 (8) | 0.038 (5) | 0.001 (5) | 0.009 (5) | −0.003 (5) |
| C13 | 0.062 (7) | 0.042 (6) | 0.056 (7) | −0.003 (5) | 0.004 (5) | 0.001 (6) |
| C14 | 0.054 (3) | 0.059 (10) | 0.051 (3) | 0.001 (5) | 0.002 (3) | 0.006 (4) |
Geometric parameters (Å, º) top
| Cu1—O1 | 1.934 (2) | C5—C5i | 1.426 (5) |
| Cu1—O1i | 1.934 (2) | C6—C7 | 1.396 (5) |
| Cu1—N1 | 2.028 (3) | C6—C10 | 1.442 (4) |
| Cu1—N1i | 2.028 (3) | C7—C8 | 1.368 (5) |
| Cu1—O1W | 2.279 (4) | C7—H7 | 0.9300 |
| O1—C1 | 1.265 (4) | C8—C9 | 1.394 (5) |
| O2—C1 | 1.235 (5) | C8—H8 | 0.9300 |
| O1W—H1W1 | 0.84 (1) | C9—H9 | 0.9300 |
| O1W—H1W2 | 0.85 (1) | C10—C10i | 1.388 (6) |
| O2W—H2W1 | 0.85 (1) | C11—C11i | 1.374 (6) |
| O2W—H2W2 | 0.86 (1) | C11—H11 | 0.9300 |
| N1—C9 | 1.324 (4) | O5—C12 | 1.261 (10) |
| N1—C5 | 1.353 (3) | N3—C12 | 1.338 (9) |
| N2—C11 | 1.156 (5) | N3—C13 | 1.442 (9) |
| N2—C10 | 1.397 (4) | N3—C14 | 1.448 (6) |
| C1—C2 | 1.516 (4) | C12—H12 | 0.9300 |
| C2—C3 | 1.378 (5) | C13—H13A | 0.9600 |
| C2—C4ii | 1.391 (5) | C13—H13B | 0.9600 |
| C3—C4 | 1.387 (4) | C13—H13C | 0.9600 |
| C3—H3 | 0.9300 | C14—H14A | 0.9600 |
| C4—C2ii | 1.391 (5) | C14—H14B | 0.9600 |
| C4—H4 | 0.9300 | C14—H14C | 0.9600 |
| C5—C6 | 1.401 (4) | | |
| | | |
| O1—Cu1—O1i | 90.5 (2) | C8—C7—C6 | 119.2 (3) |
| O1—Cu1—O1W | 91.8 (2) | C8—C7—H7 | 120.4 |
| O1—Cu1—N1 | 94.1 (1) | C6—C7—H7 | 120.4 |
| O1—Cu1—N1i | 174.7 (1) | C7—C8—C9 | 119.6 (3) |
| O1W—Cu1—N1 | 90.6 (1) | C7—C8—H8 | 120.2 |
| O1W—Cu1—N1i | 90.6 (1) | C9—C8—H8 | 120.2 |
| N1—Cu1—N1i | 81.1 (1) | N1—C9—C8 | 122.3 (3) |
| C1—O1—Cu1 | 127.1 (3) | N1—C9—H9 | 118.8 |
| Cu1—O1W—H1W1 | 125 (4) | C8—C9—H9 | 118.8 |
| Cu1—O1W—H1W2 | 125 (3) | C10i—C10—N2 | 119.9 (2) |
| H1W1—O1W—H1W2 | 110 (2) | C10i—C10—C6 | 120.97 (17) |
| H2W1—O2W—H2W2 | 109 (2) | N2—C10—C6 | 119.1 (3) |
| C9—N1—C5 | 118.7 (3) | N2—C11—C11i | 127.5 (2) |
| C9—N1—Cu1 | 128.4 (2) | N2—C11—H11 | 116.2 |
| C5—N1—Cu1 | 112.88 (19) | C11i—C11—H11 | 116.2 |
| C11—N2—C10 | 112.5 (4) | C12—N3—C13 | 120.3 (5) |
| O2—C1—O1 | 126.3 (3) | C12—N3—C14 | 121.3 (11) |
| O2—C1—C2 | 118.9 (3) | C13—N3—C14 | 118.4 (11) |
| O1—C1—C2 | 114.8 (3) | O5—C12—N3 | 121.8 (15) |
| C3—C2—C4ii | 119.4 (3) | O5—C12—H12 | 119.1 |
| C3—C2—C1 | 120.6 (3) | N3—C12—H12 | 119.1 |
| C4ii—C2—C1 | 119.9 (3) | N3—C13—H13A | 109.5 |
| C2—C3—C4 | 120.5 (3) | N3—C13—H13B | 109.5 |
| C2—C3—H3 | 119.7 | H13A—C13—H13B | 109.5 |
| C4—C3—H3 | 119.7 | N3—C13—H13C | 109.5 |
| C3—C4—C2ii | 120.1 (3) | H13A—C13—H13C | 109.5 |
| C3—C4—H4 | 120.0 | H13B—C13—H13C | 109.5 |
| C2ii—C4—H4 | 120.0 | N3—C14—H14A | 109.5 |
| N1—C5—C6 | 122.4 (3) | N3—C14—H14B | 109.5 |
| N1—C5—C5i | 116.57 (15) | H14A—C14—H14B | 109.5 |
| C6—C5—C5i | 121.04 (18) | N3—C14—H14C | 109.5 |
| C7—C6—C5 | 117.8 (3) | H14A—C14—H14C | 109.5 |
| C7—C6—C10 | 124.1 (3) | H14B—C14—H14C | 109.5 |
| C5—C6—C10 | 118.0 (3) | | |
| | | |
| O1i—Cu1—O1—C1 | −95.8 (3) | Cu1—N1—C5—C5i | 0.8 (2) |
| N1—Cu1—O1—C1 | 81.6 (3) | N1—C5—C6—C7 | −0.9 (5) |
| O1W—Cu1—O1—C1 | 172.4 (3) | C5i—C5—C6—C7 | −178.8 (3) |
| O1—Cu1—N1—C9 | 4.8 (3) | N1—C5—C6—C10 | 176.1 (3) |
| N1i—Cu1—N1—C9 | −177.6 (3) | C5i—C5—C6—C10 | −1.8 (4) |
| O1W—Cu1—N1—C9 | −87.1 (3) | C5—C6—C7—C8 | 1.6 (6) |
| O1—Cu1—N1—C5 | −178.6 (2) | C10—C6—C7—C8 | −175.2 (4) |
| N1i—Cu1—N1—C5 | −0.9 (3) | C6—C7—C8—C9 | −1.2 (7) |
| O1W—Cu1—N1—C5 | 89.6 (2) | C5—N1—C9—C8 | 0.6 (6) |
| Cu1—O1—C1—O2 | −14.0 (5) | Cu1—N1—C9—C8 | 177.1 (3) |
| Cu1—O1—C1—C2 | 167.1 (2) | C7—C8—C9—N1 | 0.1 (7) |
| O2—C1—C2—C3 | −177.9 (3) | C11—N2—C10—C10i | −2.2 (4) |
| O1—C1—C2—C3 | 1.1 (4) | C11—N2—C10—C6 | 176.0 (3) |
| O2—C1—C2—C4ii | 3.4 (5) | C7—C6—C10—C10i | 178.6 (3) |
| O1—C1—C2—C4ii | −177.6 (3) | C5—C6—C10—C10i | 1.8 (4) |
| C4ii—C2—C3—C4 | 0.2 (5) | C7—C6—C10—N2 | 0.4 (5) |
| C1—C2—C3—C4 | −178.5 (3) | C5—C6—C10—N2 | −176.4 (3) |
| C2—C3—C4—C2ii | −0.2 (5) | C10—N2—C11—C11i | 2.4 (4) |
| C9—N1—C5—C6 | −0.2 (5) | C13—N3—C12—O5 | 0.3 (3) |
| Cu1—N1—C5—C6 | −177.2 (2) | C14—N3—C12—O5 | 179.6 (3) |
| C9—N1—C5—C5i | 177.8 (3) | | |
| Symmetry codes: (i) x, −y+1/2, z; (ii) −x+1, −y+1, −z+2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1W—H1W1···O5 | 0.84 (1) | 2.01 (1) | 2.708 (7) | 140 (1) |
| O1W—H1W2···O2W | 0.85 (1) | 1.87 (1) | 2.716 (7) | 175 (3) |
| O2W—H2W1···O2iii | 0.85 (1) | 1.77 (5) | 2.61 (4) | 167 (9) |
| Symmetry code: (iii) x+1, y, z. |
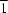 site symmetry. This dianion serves as a spacer to connect adjacent square-pyramidal metal atoms into a helical chain that propagates along the b axis of the monoclinic unit cell. The chain motif is consolidated into a layer structure by hydrogen bonds involving the coordinated and uncoordinated water molecules. The Cu atom and the coordinated water molecule lie on a mirror plane, while the N-heterocycle, the disordered uncoordinated water molecule and disordered dimethylformamide solvent molecule lie about a mirror plane.
site symmetry. This dianion serves as a spacer to connect adjacent square-pyramidal metal atoms into a helical chain that propagates along the b axis of the monoclinic unit cell. The chain motif is consolidated into a layer structure by hydrogen bonds involving the coordinated and uncoordinated water molecules. The Cu atom and the coordinated water molecule lie on a mirror plane, while the N-heterocycle, the disordered uncoordinated water molecule and disordered dimethylformamide solvent molecule lie about a mirror plane.
 journal menu
journal menu











 Alert level A
Alert level A
 Alert level B
Alert level B
 Alert level C
Alert level C
 Alert level G
Alert level G



