
In the title compounds, C
7H
8NO
2+·NO
3−, (I), C
7H
8NO
2+·ClO
4−·H
2O, (II), and 2C
7H
8NO
2+·SO
42−, (III), the carboxyl planes of the 4-carboxyphenylammonium cations are twisted from the aromatic plane. A homonuclear
C(8) hydrogen-bonding motif of 4-carboxyphenylammonium cations is observed in both (I) and (II), leading to `head-to-tail' layers. The cations in (III) form carboxyl group dimers, making a graph-set motif of
R22(8). In all the structures, anions connect the cationic layers and an infinite chain running along the
c axis is observed, having the
C22(6) graph-set motif. Interestingly, in (II), the anions are connected through weak hydrogen bonds involving the water molecules, leading to a graph-set motif of
R44(12). Alternate hydrophobic and hydrophilic layers are observed in all three compounds as a result of the column-like arrangement of the aromatic rings of the cations and the anions. Furthermore, in (I), head-to-tail N—H

O interactions and interactions linking the cations and anions form an
R64(16) hydrogen-bonding motif, resulting in a pseudo-inversion centre at (
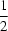
,
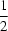
, 0).
Supporting information
CCDC references: 625694; 625695; 625696
The title compounds, (I), (II) and (III), were crystallized from aqueous solutions containing p-aminobenzoic acid with nitric acid, perchloric acid and sulfuric acid in the stochiometric ratios 1:1, 1:1 and 1:2, respectively, at room temperature, by the technique of slow evaporation.
The water H atoms in (II) were located and refined isotropically. All other H atoms were positioned geometrically and refined using a riding model, with C—H = 0.93 Å, O—H = 0.82 Å and N—H = 0.89 Å and Uiso(H) = 1.2–1.5Ueq(parent atom). In structure (I), the hydroxy group O1B is disordered over two positions with major and minor site occupancies of 0.75 and 1/4, respectively.
For all compounds, data collection: CAD-4 EXPRESS (Enraf–Nonius, 1994); cell refinement: CAD-4 EXPRESS; data reduction: XCAD4 (Harms & Wocadlo, 1995); program(s) used to solve structure: SHELXTL/PC (Bruker, 2000); program(s) used to refine structure: SHELXTL/PC. Molecular graphics: SHELXTL/PC and PLATON (Spek, 2003) for (I); SHELXTL/PC, Mercury (Version 1.4.1, Macrae et al., 2006) and PLATON (Spek, 2003) for (II), (III). For all compounds, software used to prepare material for publication: SHELXL97 (Sheldrick, 1997).
Crystal data top
| C7H8NO2+·NO3− | F(000) = 416 |
| Mr = 200.15 | Dx = 1.497 Mg m−3
Dm = 1.49 Mg m−3
Dm measured by flotation using a liquid mixture of xylene and bromoform |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 8.8154 (9) Å | θ = 9.9–14.4° |
| b = 15.0516 (3) Å | µ = 0.13 mm−1 |
| c = 6.6950 (6) Å | T = 293 K |
| β = 90.933 (12)° | Block, light pink |
| V = 888.14 (14) Å3 | 0.22 × 0.19 × 0.17 mm |
| Z = 4 | |
Data collection top
Nonius MACH3 sealed tube
diffractometer | Rint = 0.016 |
| Radiation source: fine-focus sealed tube | θmax = 25.0°, θmin = 2.3° |
| Graphite monochromator | h = −10→10 |
| ω–2θ scans | k = −1→17 |
| 1832 measured reflections | l = 0→7 |
| 1549 independent reflections | 3 standard reflections every 60 min |
| 1264 reflections with I > 2σ(I) | intensity decay: none |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.047 | H-atom parameters constrained |
| wR(F2) = 0.134 | w = 1/[σ2(Fo2) + (0.0712P)2 + 0.3551P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 1549 reflections | Δρmax = 0.29 e Å−3 |
| 139 parameters | Δρmin = −0.21 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.047 (6) |
Crystal data top
| C7H8NO2+·NO3− | V = 888.14 (14) Å3 |
| Mr = 200.15 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 8.8154 (9) Å | µ = 0.13 mm−1 |
| b = 15.0516 (3) Å | T = 293 K |
| c = 6.6950 (6) Å | 0.22 × 0.19 × 0.17 mm |
| β = 90.933 (12)° | |
Data collection top
Nonius MACH3 sealed tube
diffractometer | Rint = 0.016 |
| 1832 measured reflections | 3 standard reflections every 60 min |
| 1549 independent reflections | intensity decay: none |
| 1264 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.047 | 0 restraints |
| wR(F2) = 0.134 | H-atom parameters constrained |
| S = 1.08 | Δρmax = 0.29 e Å−3 |
| 1549 reflections | Δρmin = −0.21 e Å−3 |
| 139 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| C1 | 1.0614 (2) | 0.26541 (13) | 0.7285 (3) | 0.0402 (5) | |
| C2 | 0.9128 (2) | 0.23648 (13) | 0.7434 (3) | 0.0415 (5) | |
| H2 | 0.8916 | 0.1760 | 0.7483 | 0.050* | |
| C3 | 0.7965 (2) | 0.29739 (14) | 0.7508 (3) | 0.0430 (5) | |
| H3 | 0.6963 | 0.2785 | 0.7589 | 0.052* | |
| C4 | 0.8308 (2) | 0.38687 (12) | 0.7461 (3) | 0.0365 (5) | |
| C5 | 0.9772 (2) | 0.41687 (13) | 0.7335 (3) | 0.0452 (5) | |
| H5 | 0.9980 | 0.4774 | 0.7318 | 0.054* | |
| C6 | 1.0933 (2) | 0.35541 (14) | 0.7234 (4) | 0.0480 (6) | |
| H6 | 1.1932 | 0.3746 | 0.7131 | 0.058* | |
| C11 | 1.1830 (2) | 0.19752 (15) | 0.7135 (4) | 0.0504 (6) | |
| O1A | 1.16056 (18) | 0.11868 (10) | 0.7343 (3) | 0.0564 (5) | |
| O1B | 1.3149 (3) | 0.2309 (2) | 0.6546 (4) | 0.0552 (8) | 0.75 |
| H1B | 1.3780 | 0.1910 | 0.6486 | 0.083* | 0.75 |
| O1B' | 1.3167 (9) | 0.2319 (6) | 0.792 (3) | 0.094 (4) | 0.25 |
| H1B' | 1.3832 | 0.1939 | 0.7884 | 0.142* | 0.25 |
| N4 | 0.70517 (18) | 0.45084 (11) | 0.7523 (3) | 0.0415 (5) | |
| H4A | 0.7419 | 0.5058 | 0.7457 | 0.062* | |
| H4B | 0.6552 | 0.4441 | 0.8658 | 0.062* | |
| H4C | 0.6423 | 0.4413 | 0.6492 | 0.062* | |
| N1 | 0.59434 (19) | 0.41968 (12) | 0.2482 (3) | 0.0486 (5) | |
| O1 | 0.5333 (2) | 0.39844 (15) | 0.4074 (3) | 0.0763 (6) | |
| O2 | 0.5345 (2) | 0.39385 (13) | 0.0872 (3) | 0.0662 (6) | |
| O3 | 0.7100 (2) | 0.46391 (14) | 0.2473 (4) | 0.0836 (7) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0393 (11) | 0.0329 (10) | 0.0483 (12) | 0.0007 (8) | 0.0006 (8) | 0.0009 (9) |
| C2 | 0.0432 (11) | 0.0277 (10) | 0.0536 (12) | −0.0027 (8) | 0.0023 (9) | 0.0009 (9) |
| C3 | 0.0344 (10) | 0.0370 (11) | 0.0575 (13) | −0.0058 (8) | 0.0022 (9) | 0.0009 (9) |
| C4 | 0.0346 (10) | 0.0324 (10) | 0.0427 (10) | 0.0015 (8) | 0.0019 (8) | 0.0001 (8) |
| C5 | 0.0377 (11) | 0.0285 (10) | 0.0695 (14) | −0.0031 (9) | 0.0019 (9) | 0.0011 (9) |
| C6 | 0.0321 (10) | 0.0368 (11) | 0.0752 (15) | −0.0037 (8) | 0.0014 (9) | 0.0008 (10) |
| C11 | 0.0416 (12) | 0.0414 (12) | 0.0683 (15) | 0.0032 (9) | 0.0008 (10) | −0.0001 (10) |
| O1A | 0.0524 (9) | 0.0329 (8) | 0.0842 (12) | 0.0071 (7) | 0.0052 (8) | 0.0022 (7) |
| O1B | 0.0419 (13) | 0.0481 (15) | 0.0759 (17) | 0.0088 (10) | 0.0132 (13) | 0.0070 (14) |
| O1B' | 0.031 (4) | 0.036 (4) | 0.216 (14) | 0.009 (3) | 0.006 (7) | −0.008 (7) |
| N4 | 0.0349 (9) | 0.0354 (9) | 0.0543 (10) | 0.0017 (7) | 0.0036 (7) | 0.0001 (7) |
| N1 | 0.0321 (9) | 0.0389 (10) | 0.0749 (14) | 0.0038 (8) | 0.0010 (9) | −0.0011 (9) |
| O1 | 0.0634 (12) | 0.1024 (16) | 0.0629 (11) | −0.0225 (10) | −0.0017 (9) | 0.0012 (10) |
| O2 | 0.0552 (10) | 0.0820 (13) | 0.0615 (11) | −0.0216 (9) | 0.0046 (8) | −0.0048 (9) |
| O3 | 0.0521 (11) | 0.0696 (13) | 0.1290 (19) | −0.0285 (9) | 0.0008 (11) | −0.0035 (12) |
Geometric parameters (Å, º) top
| C1—C6 | 1.384 (3) | C11—O1A | 1.211 (3) |
| C1—C2 | 1.386 (3) | C11—O1B | 1.333 (3) |
| C1—C11 | 1.486 (3) | C11—O1B' | 1.383 (10) |
| C2—C3 | 1.377 (3) | O1B—H1B | 0.8200 |
| C2—H2 | 0.9300 | O1B'—H1B' | 0.8200 |
| C3—C4 | 1.381 (3) | N4—H4A | 0.8900 |
| C3—H3 | 0.9300 | N4—H4B | 0.8900 |
| C4—C5 | 1.371 (3) | N4—H4C | 0.8900 |
| C4—N4 | 1.468 (2) | N1—O3 | 1.217 (2) |
| C5—C6 | 1.382 (3) | N1—O1 | 1.244 (3) |
| C5—H5 | 0.9300 | N1—O2 | 1.254 (3) |
| C6—H6 | 0.9300 | | |
| | | |
| C6—C1—C2 | 120.16 (19) | O1A—C11—O1B | 123.3 (2) |
| C6—C1—C11 | 121.61 (18) | O1A—C11—O1B' | 117.6 (5) |
| C2—C1—C11 | 118.22 (18) | O1B—C11—O1B' | 39.5 (6) |
| C3—C2—C1 | 119.91 (18) | O1A—C11—C1 | 123.1 (2) |
| C3—C2—H2 | 120.0 | O1B—C11—C1 | 113.2 (2) |
| C1—C2—H2 | 120.0 | O1B'—C11—C1 | 109.2 (4) |
| C2—C3—C4 | 119.04 (18) | C11—O1B—H1B | 109.5 |
| C2—C3—H3 | 120.5 | C11—O1B'—H1B' | 109.5 |
| C4—C3—H3 | 120.5 | C4—N4—H4A | 109.5 |
| C5—C4—C3 | 121.94 (18) | C4—N4—H4B | 109.5 |
| C5—C4—N4 | 119.80 (17) | H4A—N4—H4B | 109.5 |
| C3—C4—N4 | 118.26 (17) | C4—N4—H4C | 109.5 |
| C4—C5—C6 | 118.77 (18) | H4A—N4—H4C | 109.5 |
| C4—C5—H5 | 120.6 | H4B—N4—H4C | 109.5 |
| C6—C5—H5 | 120.6 | O3—N1—O1 | 121.3 (2) |
| C5—C6—C1 | 120.17 (19) | O3—N1—O2 | 120.4 (2) |
| C5—C6—H6 | 119.9 | O1—N1—O2 | 118.35 (18) |
| C1—C6—H6 | 119.9 | | |
| | | |
| C6—C1—C2—C3 | 0.7 (3) | C2—C1—C6—C5 | 0.1 (3) |
| C11—C1—C2—C3 | −177.81 (19) | C11—C1—C6—C5 | 178.6 (2) |
| C1—C2—C3—C4 | −0.9 (3) | C6—C1—C11—O1A | 173.6 (2) |
| C2—C3—C4—C5 | 0.2 (3) | C2—C1—C11—O1A | −7.9 (3) |
| C2—C3—C4—N4 | 179.44 (18) | C6—C1—C11—O1B | −13.0 (3) |
| C3—C4—C5—C6 | 0.6 (3) | C2—C1—C11—O1B | 165.5 (2) |
| N4—C4—C5—C6 | −178.57 (19) | C6—C1—C11—O1B' | 29.3 (8) |
| C4—C5—C6—C1 | −0.8 (3) | C2—C1—C11—O1B' | −152.2 (7) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1B—H1B···O1i | 0.82 | 2.57 | 3.202 (3) | 135 |
| O1B′—H1B′···O2i | 0.82 | 2.32 | 3.038 (10) | 146 |
| N4—H4A···O1Aii | 0.89 | 1.91 | 2.791 (2) | 171 |
| N4—H4B···O2iii | 0.89 | 1.99 | 2.853 (2) | 163 |
| N4—H4C···O1 | 0.89 | 1.98 | 2.852 (3) | 167 |
| Symmetry codes: (i) x+1, −y+1/2, z+1/2; (ii) −x+2, y+1/2, −z+3/2; (iii) x, y, z+1. |
Crystal data top
| C7H8NO2+·ClO4−·H2O | F(000) = 528 |
| Mr = 255.61 | Dx = 1.622 Mg m−3
Dm = 1.61 Mg m−3
Dm measured by flotation technique using a liquid mixture of carbon tetrachloride and bromoform |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 7.5492 (7) Å | θ = 11.5–14.3° |
| b = 19.3871 (5) Å | µ = 0.39 mm−1 |
| c = 7.2974 (3) Å | T = 293 K |
| β = 101.524 (5)° | Block, colourless |
| V = 1046.42 (12) Å3 | 0.23 × 0.20 × 0.18 mm |
| Z = 4 | |
Data collection top
Nonius MACH3 sealed tube
diffractometer | 1587 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.029 |
| Graphite monochromator | θmax = 25.0°, θmin = 2.1° |
| ω–2θ scans | h = −8→8 |
Absorption correction: ψ scan
(North et al., 1968) | k = −23→23 |
| Tmin = 0.911, Tmax = 0.994 | l = 0→8 |
| 3032 measured reflections | 3 standard reflections every 60 min |
| 1833 independent reflections | intensity decay: none |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.120 | w = 1/[σ2(Fo2) + (0.0592P)2 + 0.8272P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 1833 reflections | Δρmax = 0.52 e Å−3 |
| 156 parameters | Δρmin = −0.33 e Å−3 |
| 2 restraints | Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.049 (4) |
Crystal data top
| C7H8NO2+·ClO4−·H2O | V = 1046.42 (12) Å3 |
| Mr = 255.61 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 7.5492 (7) Å | µ = 0.39 mm−1 |
| b = 19.3871 (5) Å | T = 293 K |
| c = 7.2974 (3) Å | 0.23 × 0.20 × 0.18 mm |
| β = 101.524 (5)° | |
Data collection top
Nonius MACH3 sealed tube
diffractometer | 1587 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(North et al., 1968) | Rint = 0.029 |
| Tmin = 0.911, Tmax = 0.994 | 3 standard reflections every 60 min |
| 3032 measured reflections | intensity decay: none |
| 1833 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.042 | 2 restraints |
| wR(F2) = 0.120 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | Δρmax = 0.52 e Å−3 |
| 1833 reflections | Δρmin = −0.33 e Å−3 |
| 156 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.3520 (3) | 0.26926 (11) | 0.9953 (3) | 0.0303 (5) | |
| C2 | 0.3959 (3) | 0.19965 (12) | 1.0169 (3) | 0.0356 (6) | |
| H2 | 0.5120 | 0.1865 | 1.0750 | 0.043* | |
| C3 | 0.2671 (3) | 0.15006 (12) | 0.9523 (3) | 0.0363 (6) | |
| H3 | 0.2960 | 0.1034 | 0.9651 | 0.044* | |
| C4 | 0.0950 (3) | 0.17067 (11) | 0.8685 (3) | 0.0311 (5) | |
| C5 | 0.0483 (3) | 0.23973 (12) | 0.8465 (3) | 0.0354 (5) | |
| H5 | −0.0684 | 0.2528 | 0.7901 | 0.042* | |
| C6 | 0.1785 (3) | 0.28871 (12) | 0.9098 (3) | 0.0347 (5) | |
| H6 | 0.1496 | 0.3353 | 0.8951 | 0.042* | |
| N4 | −0.0423 (3) | 0.11817 (10) | 0.8007 (3) | 0.0371 (5) | |
| H4A | −0.1456 | 0.1387 | 0.7488 | 0.056* | |
| H4B | −0.0596 | 0.0922 | 0.8961 | 0.056* | |
| H4C | −0.0050 | 0.0918 | 0.7161 | 0.056* | |
| C11 | 0.4921 (3) | 0.32139 (12) | 1.0682 (3) | 0.0340 (5) | |
| O1A | 0.6400 (2) | 0.30696 (9) | 1.1586 (3) | 0.0488 (5) | |
| O1B | 0.4390 (2) | 0.38523 (9) | 1.0268 (3) | 0.0491 (5) | |
| H1B | 0.5171 | 0.4121 | 1.0785 | 0.074* | |
| Cl1 | 0.80735 (8) | 0.44858 (3) | 0.75074 (8) | 0.0401 (3) | |
| O1 | 0.8161 (3) | 0.51293 (12) | 0.8533 (4) | 0.0712 (7) | |
| O2 | 0.9551 (4) | 0.44862 (14) | 0.6567 (4) | 0.0793 (8) | |
| O3 | 0.8212 (4) | 0.39423 (12) | 0.8810 (4) | 0.0755 (7) | |
| O4 | 0.6381 (4) | 0.44810 (16) | 0.6250 (4) | 0.0877 (9) | |
| O1W | 0.3392 (3) | 0.52034 (10) | 0.7949 (3) | 0.0506 (5) | |
| H1W | 0.386 (5) | 0.502 (2) | 0.719 (4) | 0.080 (14)* | |
| H2W | 0.295 (6) | 0.491 (2) | 0.850 (6) | 0.109 (17)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0281 (11) | 0.0315 (11) | 0.0306 (11) | 0.0003 (9) | 0.0041 (9) | 0.0004 (9) |
| C2 | 0.0273 (11) | 0.0353 (12) | 0.0403 (13) | 0.0048 (9) | −0.0026 (9) | 0.0024 (10) |
| C3 | 0.0356 (12) | 0.0275 (11) | 0.0424 (13) | 0.0039 (9) | −0.0004 (10) | 0.0014 (10) |
| C4 | 0.0307 (11) | 0.0317 (11) | 0.0291 (11) | −0.0029 (9) | 0.0016 (9) | −0.0007 (9) |
| C5 | 0.0258 (11) | 0.0357 (12) | 0.0409 (13) | 0.0043 (9) | −0.0023 (9) | 0.0012 (10) |
| C6 | 0.0316 (12) | 0.0280 (11) | 0.0426 (13) | 0.0037 (9) | 0.0029 (10) | 0.0024 (10) |
| N4 | 0.0325 (10) | 0.0342 (10) | 0.0407 (11) | −0.0025 (8) | −0.0022 (8) | −0.0012 (9) |
| C11 | 0.0311 (12) | 0.0333 (12) | 0.0367 (12) | −0.0007 (9) | 0.0046 (9) | 0.0019 (10) |
| O1A | 0.0328 (9) | 0.0386 (10) | 0.0669 (13) | −0.0042 (7) | −0.0093 (8) | 0.0064 (9) |
| O1B | 0.0392 (10) | 0.0303 (9) | 0.0702 (13) | −0.0012 (7) | −0.0077 (9) | 0.0000 (9) |
| Cl1 | 0.0412 (4) | 0.0394 (4) | 0.0390 (4) | −0.0017 (2) | 0.0067 (3) | 0.0057 (2) |
| O1 | 0.0846 (17) | 0.0518 (13) | 0.0805 (16) | −0.0033 (11) | 0.0241 (13) | −0.0134 (12) |
| O2 | 0.0811 (17) | 0.102 (2) | 0.0657 (15) | −0.0181 (14) | 0.0399 (13) | −0.0167 (14) |
| O3 | 0.0924 (18) | 0.0600 (14) | 0.0791 (16) | 0.0131 (12) | 0.0289 (14) | 0.0306 (12) |
| O4 | 0.0640 (15) | 0.107 (2) | 0.0786 (17) | −0.0162 (14) | −0.0181 (13) | 0.0087 (15) |
| O1W | 0.0485 (11) | 0.0396 (11) | 0.0606 (13) | −0.0032 (9) | 0.0034 (10) | −0.0022 (10) |
Geometric parameters (Å, º) top
| C1—C6 | 1.386 (3) | N4—H4A | 0.8900 |
| C1—C2 | 1.391 (3) | N4—H4B | 0.8900 |
| C1—C11 | 1.483 (3) | N4—H4C | 0.8900 |
| C2—C3 | 1.381 (3) | C11—O1A | 1.211 (3) |
| C2—H2 | 0.9300 | C11—O1B | 1.317 (3) |
| C3—C4 | 1.380 (3) | O1B—H1B | 0.8200 |
| C3—H3 | 0.9300 | Cl1—O3 | 1.409 (2) |
| C4—C5 | 1.386 (3) | Cl1—O4 | 1.416 (2) |
| C4—N4 | 1.466 (3) | Cl1—O2 | 1.421 (2) |
| C5—C6 | 1.378 (3) | Cl1—O1 | 1.449 (2) |
| C5—H5 | 0.9300 | O1W—H1W | 0.794 (19) |
| C6—H6 | 0.9300 | O1W—H2W | 0.81 (2) |
| | | |
| C6—C1—C2 | 119.8 (2) | C4—N4—H4A | 109.5 |
| C6—C1—C11 | 121.2 (2) | C4—N4—H4B | 109.5 |
| C2—C1—C11 | 118.9 (2) | H4A—N4—H4B | 109.5 |
| C3—C2—C1 | 120.1 (2) | C4—N4—H4C | 109.5 |
| C3—C2—H2 | 120.0 | H4A—N4—H4C | 109.5 |
| C1—C2—H2 | 120.0 | H4B—N4—H4C | 109.5 |
| C4—C3—C2 | 119.1 (2) | O1A—C11—O1B | 123.2 (2) |
| C4—C3—H3 | 120.5 | O1A—C11—C1 | 123.5 (2) |
| C2—C3—H3 | 120.5 | O1B—C11—C1 | 113.30 (19) |
| C3—C4—C5 | 121.7 (2) | C11—O1B—H1B | 109.5 |
| C3—C4—N4 | 119.2 (2) | O3—Cl1—O4 | 111.62 (17) |
| C5—C4—N4 | 119.06 (19) | O3—Cl1—O2 | 111.48 (16) |
| C6—C5—C4 | 118.6 (2) | O4—Cl1—O2 | 112.36 (18) |
| C6—C5—H5 | 120.7 | O3—Cl1—O1 | 107.81 (16) |
| C4—C5—H5 | 120.7 | O4—Cl1—O1 | 106.41 (17) |
| C5—C6—C1 | 120.7 (2) | O2—Cl1—O1 | 106.80 (15) |
| C5—C6—H6 | 119.7 | H1W—O1W—H2W | 109 (5) |
| C1—C6—H6 | 119.7 | | |
| | | |
| C6—C1—C2—C3 | 0.4 (4) | C4—C5—C6—C1 | −0.6 (4) |
| C11—C1—C2—C3 | 179.2 (2) | C2—C1—C6—C5 | 0.3 (4) |
| C1—C2—C3—C4 | −0.7 (4) | C11—C1—C6—C5 | −178.5 (2) |
| C2—C3—C4—C5 | 0.4 (4) | C6—C1—C11—O1A | 173.0 (2) |
| C2—C3—C4—N4 | −179.6 (2) | C2—C1—C11—O1A | −5.8 (4) |
| C3—C4—C5—C6 | 0.2 (4) | C6—C1—C11—O1B | −6.1 (3) |
| N4—C4—C5—C6 | −179.7 (2) | C2—C1—C11—O1B | 175.1 (2) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H4A···O1Ai | 0.89 | 1.93 | 2.818 (3) | 173 |
| N4—H4B···O2ii | 0.89 | 2.04 | 2.906 (3) | 164 |
| N4—H4C···O1iii | 0.89 | 2.22 | 3.018 (3) | 150 |
| N4—H4C···O3i | 0.89 | 2.55 | 3.039 (3) | 116 |
| O1B—H1B···O1Wiv | 0.82 | 1.83 | 2.641 (3) | 171 |
| O1W—H1W···O4 | 0.79 (2) | 2.39 (3) | 3.116 (4) | 152 (4) |
| O1W—H2W···O1iv | 0.81 (2) | 2.47 (4) | 3.096 (4) | 135 (4) |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x−1, −y+1/2, z+1/2; (iii) −x+1, y−1/2, −z+3/2; (iv) −x+1, −y+1, −z+2. |
Crystal data top
| 2C7H8NO2+·SO42− | F(000) = 776 |
| Mr = 372.35 | Dx = 1.566 Mg m−3
Dm = 1.56 Mg m−3
Dm measured by flotation technique using a liquid mixture of carbon xylene and bromoform |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 25 reflections |
| a = 18.7094 (8) Å | θ = 10.2–13.2° |
| b = 14.0562 (4) Å | µ = 0.25 mm−1 |
| c = 6.0551 (5) Å | T = 293 K |
| β = 97.272 (5)° | Block, colourless |
| V = 1579.51 (15) Å3 | 0.26 × 0.22 × 0.18 mm |
| Z = 4 | |
Data collection top
Nonius MACH3 sealed tube
diffractometer | 2014 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.045 |
| Graphite monochromator | θmax = 25.0°, θmin = 2.2° |
| ω–2θ scans | h = −22→22 |
Absorption correction: ψ scan
(North et al., 1968) | k = −1→16 |
| Tmin = 0.953, Tmax = 0.997 | l = 0→7 |
| 3323 measured reflections | 3 standard reflections every 60 min |
| 2763 independent reflections | intensity decay: none |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.060 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.181 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.1287P)2 + 0.1992P]
where P = (Fo2 + 2Fc2)/3 |
| 2763 reflections | (Δ/σ)max < 0.001 |
| 230 parameters | Δρmax = 0.84 e Å−3 |
| 0 restraints | Δρmin = −0.69 e Å−3 |
Crystal data top
| 2C7H8NO2+·SO42− | V = 1579.51 (15) Å3 |
| Mr = 372.35 | Z = 4 |
| Monoclinic, P21/c | Mo Kα radiation |
| a = 18.7094 (8) Å | µ = 0.25 mm−1 |
| b = 14.0562 (4) Å | T = 293 K |
| c = 6.0551 (5) Å | 0.26 × 0.22 × 0.18 mm |
| β = 97.272 (5)° | |
Data collection top
Nonius MACH3 sealed tube
diffractometer | 2014 reflections with I > 2σ(I) |
Absorption correction: ψ scan
(North et al., 1968) | Rint = 0.045 |
| Tmin = 0.953, Tmax = 0.997 | 3 standard reflections every 60 min |
| 3323 measured reflections | intensity decay: none |
| 2763 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.060 | 0 restraints |
| wR(F2) = 0.181 | H-atom parameters constrained |
| S = 1.05 | Δρmax = 0.84 e Å−3 |
| 2763 reflections | Δρmin = −0.69 e Å−3 |
| 230 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C11 | 0.39211 (19) | 0.3808 (2) | 0.4043 (6) | 0.0398 (8) | |
| C111 | 0.4695 (2) | 0.3776 (3) | 0.4989 (7) | 0.0482 (10) | |
| O1A | 0.48737 (16) | 0.4072 (3) | 0.6910 (5) | 0.0739 (10) | |
| O1B | 0.51279 (16) | 0.3420 (3) | 0.3765 (6) | 0.0762 (11) | |
| H1B | 0.5527 | 0.3363 | 0.4486 | 0.114* | |
| C12 | 0.34168 (19) | 0.4139 (3) | 0.5331 (6) | 0.0424 (9) | |
| H12 | 0.3569 | 0.4387 | 0.6735 | 0.051* | |
| C13 | 0.26935 (19) | 0.4107 (3) | 0.4575 (5) | 0.0377 (8) | |
| H13 | 0.2358 | 0.4323 | 0.5467 | 0.045* | |
| C14 | 0.24710 (17) | 0.3752 (2) | 0.2474 (5) | 0.0303 (7) | |
| N14 | 0.17053 (14) | 0.3649 (2) | 0.1747 (4) | 0.0324 (6) | |
| H14A | 0.1581 | 0.3040 | 0.1824 | 0.049* | |
| H14B | 0.1607 | 0.3852 | 0.0349 | 0.049* | |
| H14C | 0.1458 | 0.3993 | 0.2624 | 0.049* | |
| C15 | 0.29666 (19) | 0.3446 (3) | 0.1117 (6) | 0.0401 (9) | |
| H15 | 0.2811 | 0.3221 | −0.0307 | 0.048* | |
| C16 | 0.3694 (2) | 0.3476 (3) | 0.1891 (6) | 0.0444 (9) | |
| H16 | 0.4031 | 0.3276 | 0.0984 | 0.053* | |
| C21 | 0.74070 (19) | 0.3673 (2) | 0.8710 (6) | 0.0364 (8) | |
| C211 | 0.66428 (19) | 0.3649 (3) | 0.7710 (6) | 0.0427 (9) | |
| O2A | 0.64839 (15) | 0.3377 (2) | 0.5768 (5) | 0.0620 (9) | |
| O2B | 0.61855 (15) | 0.3933 (3) | 0.8960 (5) | 0.0642 (9) | |
| H2B | 0.5780 | 0.3920 | 0.8263 | 0.096* | |
| C22 | 0.79444 (18) | 0.3423 (2) | 0.7468 (5) | 0.0354 (8) | |
| H22 | 0.7820 | 0.3211 | 0.6015 | 0.042* | |
| C23 | 0.86627 (18) | 0.3477 (2) | 0.8319 (5) | 0.0339 (8) | |
| H23 | 0.9020 | 0.3316 | 0.7449 | 0.041* | |
| C24 | 0.88392 (17) | 0.3777 (2) | 1.0505 (5) | 0.0271 (7) | |
| N24 | 0.95875 (14) | 0.38738 (18) | 1.1427 (4) | 0.0294 (6) | |
| H24A | 0.9687 | 0.3466 | 1.2548 | 0.044* | |
| H24B | 0.9669 | 0.4465 | 1.1924 | 0.044* | |
| H24C | 0.9867 | 0.3750 | 1.0377 | 0.044* | |
| C25 | 0.83084 (18) | 0.4025 (2) | 1.1790 (5) | 0.0345 (8) | |
| H25 | 0.8435 | 0.4227 | 1.3250 | 0.041* | |
| C26 | 0.75969 (19) | 0.3976 (3) | 1.0921 (6) | 0.0392 (8) | |
| H26 | 0.7240 | 0.4141 | 1.1790 | 0.047* | |
| S1 | 0.07132 (4) | 0.36963 (5) | 0.63025 (11) | 0.0261 (3) | |
| O1 | 0.08188 (12) | 0.44111 (15) | 0.4598 (3) | 0.0323 (6) | |
| O2 | 0.14086 (13) | 0.32125 (17) | 0.6978 (4) | 0.0393 (6) | |
| O3 | 0.05029 (13) | 0.41852 (15) | 0.8284 (3) | 0.0343 (6) | |
| O4 | 0.01590 (13) | 0.30125 (16) | 0.5442 (4) | 0.0377 (6) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C11 | 0.0322 (18) | 0.046 (2) | 0.041 (2) | −0.0016 (15) | 0.0009 (15) | −0.0001 (16) |
| C111 | 0.0333 (19) | 0.059 (2) | 0.050 (2) | −0.0007 (17) | −0.0011 (17) | −0.0067 (19) |
| O1A | 0.0402 (17) | 0.112 (3) | 0.064 (2) | 0.0064 (17) | −0.0133 (15) | −0.028 (2) |
| O1B | 0.0311 (15) | 0.115 (3) | 0.080 (2) | 0.0097 (17) | 0.0001 (15) | −0.029 (2) |
| C12 | 0.038 (2) | 0.059 (2) | 0.0288 (18) | −0.0043 (17) | −0.0008 (14) | −0.0056 (16) |
| C13 | 0.0354 (18) | 0.054 (2) | 0.0242 (17) | −0.0026 (15) | 0.0043 (13) | −0.0045 (15) |
| C14 | 0.0312 (17) | 0.0358 (17) | 0.0240 (16) | −0.0020 (13) | 0.0040 (13) | 0.0046 (13) |
| N14 | 0.0305 (14) | 0.0446 (15) | 0.0211 (13) | −0.0028 (12) | −0.0010 (11) | 0.0037 (12) |
| C15 | 0.0377 (19) | 0.056 (2) | 0.0265 (17) | −0.0038 (16) | 0.0022 (14) | −0.0087 (15) |
| C16 | 0.0359 (19) | 0.060 (2) | 0.038 (2) | −0.0001 (17) | 0.0103 (15) | −0.0073 (17) |
| C21 | 0.0332 (18) | 0.0396 (18) | 0.0354 (18) | −0.0010 (14) | 0.0001 (14) | 0.0018 (15) |
| C211 | 0.0341 (19) | 0.050 (2) | 0.043 (2) | −0.0025 (16) | 0.0006 (16) | −0.0002 (17) |
| O2A | 0.0365 (15) | 0.092 (2) | 0.0535 (18) | 0.0011 (15) | −0.0078 (13) | −0.0198 (17) |
| O2B | 0.0304 (14) | 0.103 (2) | 0.0576 (19) | 0.0070 (16) | 0.0007 (13) | −0.0120 (18) |
| C22 | 0.0376 (19) | 0.0458 (19) | 0.0215 (16) | −0.0014 (15) | −0.0011 (13) | −0.0057 (14) |
| C23 | 0.0340 (18) | 0.0437 (18) | 0.0242 (16) | 0.0001 (14) | 0.0048 (13) | −0.0045 (14) |
| C24 | 0.0290 (16) | 0.0287 (15) | 0.0236 (15) | −0.0009 (12) | 0.0032 (12) | 0.0022 (12) |
| N24 | 0.0290 (14) | 0.0361 (14) | 0.0222 (13) | 0.0002 (11) | −0.0001 (10) | 0.0017 (11) |
| C25 | 0.0400 (19) | 0.0431 (17) | 0.0204 (15) | 0.0020 (15) | 0.0038 (13) | −0.0044 (14) |
| C26 | 0.0356 (18) | 0.049 (2) | 0.0341 (19) | 0.0009 (16) | 0.0084 (15) | −0.0049 (16) |
| S1 | 0.0291 (4) | 0.0328 (4) | 0.0161 (4) | 0.0006 (3) | 0.0011 (3) | −0.0001 (3) |
| O1 | 0.0381 (13) | 0.0354 (12) | 0.0237 (12) | −0.0018 (10) | 0.0049 (9) | 0.0029 (9) |
| O2 | 0.0367 (13) | 0.0502 (13) | 0.0310 (13) | 0.0091 (11) | 0.0047 (10) | 0.0023 (11) |
| O3 | 0.0448 (14) | 0.0383 (12) | 0.0212 (11) | 0.0039 (10) | 0.0096 (10) | −0.0025 (9) |
| O4 | 0.0445 (14) | 0.0419 (13) | 0.0254 (12) | −0.0088 (11) | −0.0012 (10) | 0.0032 (10) |
Geometric parameters (Å, º) top
| C11—C12 | 1.378 (5) | C21—C211 | 1.481 (5) |
| C11—C16 | 1.399 (5) | C211—O2A | 1.237 (5) |
| C11—C111 | 1.489 (5) | C211—O2B | 1.276 (5) |
| C111—O1A | 1.241 (5) | O2B—H2B | 0.8200 |
| C111—O1B | 1.268 (5) | C22—C23 | 1.379 (5) |
| O1B—H1B | 0.8200 | C22—H22 | 0.9300 |
| C12—C13 | 1.373 (5) | C23—C24 | 1.389 (4) |
| C12—H12 | 0.9300 | C23—H23 | 0.9300 |
| C13—C14 | 1.381 (5) | C24—C25 | 1.381 (4) |
| C13—H13 | 0.9300 | C24—N24 | 1.447 (4) |
| C14—C15 | 1.383 (5) | N24—H24A | 0.8900 |
| C14—N14 | 1.452 (4) | N24—H24B | 0.8900 |
| N14—H14A | 0.8900 | N24—H24C | 0.8900 |
| N14—H14B | 0.8900 | C25—C26 | 1.370 (5) |
| N14—H14C | 0.8900 | C25—H25 | 0.9300 |
| C15—C16 | 1.383 (5) | C26—H26 | 0.9300 |
| C15—H15 | 0.9300 | S1—O4 | 1.461 (2) |
| C16—H16 | 0.9300 | S1—O1 | 1.472 (2) |
| C21—C22 | 1.375 (5) | S1—O3 | 1.478 (2) |
| C21—C26 | 1.407 (5) | S1—O2 | 1.479 (2) |
| | | |
| C12—C11—C16 | 119.4 (3) | O2A—C211—O2B | 124.2 (3) |
| C12—C11—C111 | 119.7 (3) | O2A—C211—C21 | 120.0 (3) |
| C16—C11—C111 | 120.9 (3) | O2B—C211—C21 | 115.8 (3) |
| O1A—C111—O1B | 124.4 (4) | C211—O2B—H2B | 109.5 |
| O1A—C111—C11 | 118.9 (4) | C21—C22—C23 | 121.8 (3) |
| O1B—C111—C11 | 116.7 (4) | C21—C22—H22 | 119.1 |
| C111—O1B—H1B | 109.5 | C23—C22—H22 | 119.1 |
| C13—C12—C11 | 121.1 (3) | C22—C23—C24 | 118.4 (3) |
| C13—C12—H12 | 119.4 | C22—C23—H23 | 120.8 |
| C11—C12—H12 | 119.4 | C24—C23—H23 | 120.8 |
| C12—C13—C14 | 119.2 (3) | C25—C24—C23 | 120.8 (3) |
| C12—C13—H13 | 120.4 | C25—C24—N24 | 119.3 (3) |
| C14—C13—H13 | 120.4 | C23—C24—N24 | 119.9 (3) |
| C13—C14—C15 | 120.9 (3) | C24—N24—H24A | 109.5 |
| C13—C14—N14 | 119.1 (3) | C24—N24—H24B | 109.5 |
| C15—C14—N14 | 119.9 (3) | H24A—N24—H24B | 109.5 |
| C14—N14—H14A | 109.5 | C24—N24—H24C | 109.5 |
| C14—N14—H14B | 109.5 | H24A—N24—H24C | 109.5 |
| H14A—N14—H14B | 109.5 | H24B—N24—H24C | 109.5 |
| C14—N14—H14C | 109.5 | C26—C25—C24 | 120.2 (3) |
| H14A—N14—H14C | 109.5 | C26—C25—H25 | 119.9 |
| H14B—N14—H14C | 109.5 | C24—C25—H25 | 119.9 |
| C16—C15—C14 | 119.6 (3) | C25—C26—C21 | 119.8 (3) |
| C16—C15—H15 | 120.2 | C25—C26—H26 | 120.1 |
| C14—C15—H15 | 120.2 | C21—C26—H26 | 120.1 |
| C15—C16—C11 | 119.7 (3) | O4—S1—O1 | 110.57 (12) |
| C15—C16—H16 | 120.1 | O4—S1—O3 | 110.22 (14) |
| C11—C16—H16 | 120.1 | O1—S1—O3 | 108.97 (13) |
| C22—C21—C26 | 118.9 (3) | O4—S1—O2 | 110.84 (14) |
| C22—C21—C211 | 120.4 (3) | O1—S1—O2 | 108.44 (13) |
| C26—C21—C211 | 120.7 (3) | O3—S1—O2 | 107.73 (13) |
| | | |
| C12—C11—C111—O1A | −1.8 (6) | C22—C21—C211—O2A | 2.1 (6) |
| C16—C11—C111—O1A | −179.6 (4) | C26—C21—C211—O2A | −179.6 (4) |
| C12—C11—C111—O1B | 176.5 (4) | C22—C21—C211—O2B | −177.1 (4) |
| C16—C11—C111—O1B | −1.2 (6) | C26—C21—C211—O2B | 1.2 (5) |
| C16—C11—C12—C13 | 3.0 (6) | C26—C21—C22—C23 | −1.2 (5) |
| C111—C11—C12—C13 | −174.8 (4) | C211—C21—C22—C23 | 177.1 (3) |
| C11—C12—C13—C14 | −1.0 (6) | C21—C22—C23—C24 | 1.2 (5) |
| C12—C13—C14—C15 | −1.2 (5) | C22—C23—C24—C25 | −0.7 (5) |
| C12—C13—C14—N14 | 175.2 (3) | C22—C23—C24—N24 | −177.9 (3) |
| C13—C14—C15—C16 | 1.5 (5) | C23—C24—C25—C26 | 0.2 (5) |
| N14—C14—C15—C16 | −174.9 (3) | N24—C24—C25—C26 | 177.4 (3) |
| C14—C15—C16—C11 | 0.5 (6) | C24—C25—C26—C21 | −0.2 (5) |
| C12—C11—C16—C15 | −2.7 (6) | C22—C21—C26—C25 | 0.7 (5) |
| C111—C11—C16—C15 | 175.0 (4) | C211—C21—C26—C25 | −177.7 (3) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1B—H1B···O2A | 0.82 | 1.86 | 2.670 (4) | 170 |
| N14—H14A···O2i | 0.89 | 1.79 | 2.682 (3) | 175 |
| N14—H14B···O2ii | 0.89 | 2.22 | 2.936 (3) | 137 |
| N14—H14C···O1 | 0.89 | 1.89 | 2.757 (3) | 165 |
| O2B—H2B···O1A | 0.82 | 1.80 | 2.614 (4) | 171 |
| N24—H24A···O4iii | 0.89 | 1.97 | 2.804 (3) | 156 |
| N24—H24B···O3iv | 0.89 | 1.93 | 2.741 (3) | 151 |
| N24—H24C···O3v | 0.89 | 1.94 | 2.752 (3) | 150 |
| Symmetry codes: (i) x, −y+1/2, z−1/2; (ii) x, y, z−1; (iii) x+1, y, z+1; (iv) −x+1, −y+1, −z+2; (v) x+1, y, z. |
Experimental details
| | (I) | (II) | (III) |
| Crystal data |
| Chemical formula | C7H8NO2+·NO3− | C7H8NO2+·ClO4−·H2O | 2C7H8NO2+·SO42− |
| Mr | 200.15 | 255.61 | 372.35 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 293 | 293 | 293 |
| a, b, c (Å) | 8.8154 (9), 15.0516 (3), 6.6950 (6) | 7.5492 (7), 19.3871 (5), 7.2974 (3) | 18.7094 (8), 14.0562 (4), 6.0551 (5) |
| β (°) | 90.933 (12) | 101.524 (5) | 97.272 (5) |
| V (Å3) | 888.14 (14) | 1046.42 (12) | 1579.51 (15) |
| Z | 4 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 0.13 | 0.39 | 0.25 |
| Crystal size (mm) | 0.22 × 0.19 × 0.17 | 0.23 × 0.20 × 0.18 | 0.26 × 0.22 × 0.18 |
| |
| Data collection |
| Diffractometer | Nonius MACH3 sealed tube
diffractometer | Nonius MACH3 sealed tube
diffractometer | Nonius MACH3 sealed tube
diffractometer |
| Absorption correction | – | ψ scan
(North et al., 1968) | ψ scan
(North et al., 1968) |
| Tmin, Tmax | – | 0.911, 0.994 | 0.953, 0.997 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 1832, 1549, 1264 | 3032, 1833, 1587 | 3323, 2763, 2014 |
| Rint | 0.016 | 0.029 | 0.045 |
| (sin θ/λ)max (Å−1) | 0.595 | 0.595 | 0.594 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.047, 0.134, 1.08 | 0.042, 0.120, 1.07 | 0.060, 0.181, 1.05 |
| No. of reflections | 1549 | 1833 | 2763 |
| No. of parameters | 139 | 156 | 230 |
| No. of restraints | 0 | 2 | 0 |
| H-atom treatment | H-atom parameters constrained | H atoms treated by a mixture of independent and constrained refinement | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.29, −0.21 | 0.52, −0.33 | 0.84, −0.69 |
Selected geometric parameters (Å, º) for (I) top| C4—N4 | 1.468 (2) | C11—O1B | 1.333 (3) |
| C11—O1A | 1.211 (3) | | |
| | | |
| O1A—C11—C1 | 123.1 (2) | O1B—C11—C1 | 113.2 (2) |
| | | |
| C2—C1—C11—O1A | −7.9 (3) | C6—C1—C11—O1B | −13.0 (3) |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1B—H1B···O1i | 0.82 | 2.57 | 3.202 (3) | 135 |
| O1B'—H1B'···O2i | 0.82 | 2.32 | 3.038 (10) | 146 |
| N4—H4A···O1Aii | 0.89 | 1.91 | 2.791 (2) | 171 |
| N4—H4B···O2iii | 0.89 | 1.99 | 2.853 (2) | 163 |
| N4—H4C···O1 | 0.89 | 1.98 | 2.852 (3) | 167 |
| Symmetry codes: (i) x+1, −y+1/2, z+1/2; (ii) −x+2, y+1/2, −z+3/2; (iii) x, y, z+1. |
Selected geometric parameters (Å, º) for (II) top| C4—N4 | 1.466 (3) | C11—O1B | 1.317 (3) |
| C11—O1A | 1.211 (3) | | |
| | | |
| C2—C1—C11—O1A | −5.8 (4) | C6—C1—C11—O1B | −6.1 (3) |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H4A···O1Ai | 0.89 | 1.93 | 2.818 (3) | 173 |
| N4—H4B···O2ii | 0.89 | 2.04 | 2.906 (3) | 164 |
| N4—H4C···O1iii | 0.89 | 2.22 | 3.018 (3) | 150 |
| N4—H4C···O3i | 0.89 | 2.55 | 3.039 (3) | 116 |
| O1B—H1B···O1Wiv | 0.82 | 1.83 | 2.641 (3) | 171 |
| O1W—H1W···O4 | 0.794 (19) | 2.39 (3) | 3.116 (4) | 152 (4) |
| O1W—H2W···O1iv | 0.81 (2) | 2.47 (4) | 3.096 (4) | 135 (4) |
| Symmetry codes: (i) x−1, −y+1/2, z−1/2; (ii) x−1, −y+1/2, z+1/2; (iii) −x+1, y−1/2, −z+3/2; (iv) −x+1, −y+1, −z+2. |
Selected geometric parameters (Å, º) for (III) top| C111—O1A | 1.241 (5) | C211—O2A | 1.237 (5) |
| C111—O1B | 1.268 (5) | C211—O2B | 1.276 (5) |
| C14—N14 | 1.452 (4) | C24—N24 | 1.447 (4) |
| | | |
| C12—C11—C111—O1A | −1.8 (6) | C22—C21—C211—O2A | 2.1 (6) |
| C16—C11—C111—O1A | −179.6 (4) | C26—C21—C211—O2A | −179.6 (4) |
Hydrogen-bond geometry (Å, º) for (III) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1B—H1B···O2A | 0.82 | 1.86 | 2.670 (4) | 170 |
| N14—H14A···O2i | 0.89 | 1.79 | 2.682 (3) | 175 |
| N14—H14B···O2ii | 0.89 | 2.22 | 2.936 (3) | 137 |
| N14—H14C···O1 | 0.89 | 1.89 | 2.757 (3) | 165 |
| O2B—H2B···O1A | 0.82 | 1.80 | 2.614 (4) | 171 |
| N24—H24A···O4iii | 0.89 | 1.97 | 2.804 (3) | 156 |
| N24—H24B···O3iv | 0.89 | 1.93 | 2.741 (3) | 151 |
| N24—H24C···O3v | 0.89 | 1.94 | 2.752 (3) | 150 |
| Symmetry codes: (i) x, −y+1/2, z−1/2; (ii) x, y, z−1; (iii) x+1, y, z+1; (iv) −x+1, −y+1, −z+2; (v) x+1, y, z. |
 O interactions and interactions linking the cations and anions form an R64(16) hydrogen-bonding motif, resulting in a pseudo-inversion centre at (
O interactions and interactions linking the cations and anions form an R64(16) hydrogen-bonding motif, resulting in a pseudo-inversion centre at (
 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig5thm.gif)
![[Figure 6]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig6thm.gif)
![[Figure 7]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig7thm.gif)
![[Figure 8]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig8thm.gif)
![[Figure 9]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig9thm.gif)
![[Figure 10]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig10thm.gif)
![[Figure 11]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig11thm.gif)
![[Figure 12]](http://journals.iucr.org/c/issues/2006/10/00/av3034/av3034fig12thm.gif)




p-Aminobenzoic acid (PABA) is an essential biological molecule, acting as a bacterial cofactor involved in the synthesis of folic acid. PABA is also a starting material in the manufacture of target esters, salts, folic acid, azo dyes and some other organic compounds. It is used in medicine for preparing local anesthetics and ointments. PABA has proved to be a versatile reagent for structure extension by linear hydrogen-bonding associations, through both the carboxylic acid and the amine functional groups. Considering these facts and to study the hydrogen-bonding association by different graph-set motifs, PABA was treated with three different inorganic acids, viz. nitric acid, perchloric acid and sulfuric acid, and crystals were obtained. As expected, PABA forms pronotated units with the transfer of an H atom from the inorganic acid; correspondingly, the crystal is stabilized by an extensive intermolecular hydrogen-bonding network. Crystallographic studies of PABA compounds were initiated by Pant (1965), who studied the crystal structure of 3,5-dibromo-p-aminobenzoic acid. Later, Lai & Marsh (1967) extensively studied the structure of this vitamin. The structures of 4-carboxyanilinium dihydrogenmonoarsenate monohydrate (Tordjman et al., 1988), 2,4,6-trinitrobenzoic acid 4-aminobenzoic acid monohydrate (Lynch et al., 1992), bis(p-aminobenzoic acid-N)dichlorocadmium(II) (Le Fur & Masse, 1996), 4-carboxyanilinium (2R,3R)-hydrogen tartrate monohydrate (Ziqiang et al., 2002) and bis(4-aminobenzoic acid-N)silver(I) nitrate (Wang et al., 2004) have been reported previously.
The asymmetric part of (I) contains two planar moieties, namely, a p-carboxyphenylammonium cation and a nitrate anion, nearly perpendicular to one another, with a dihedral angle of 85.89° (Fig. 1). p-Carboxyphenylammonium cations are stacked almost parallel to the (001) plane, the small angular deviation of this plane being only 3.89 (2)°. In (II), the asymmetric part of the unit cell contains a p-caboxyphenylammonium cation, a perchlorate anion and a solvent water molecule (Fig. 2). Two crystallographically independent p-carboxyphenylammonium cations and a sulfate anion constitute the asymmetric part of (III) (Fig. 3). In all three compounds, the transfer of proton(s) from the inorganic acid lead(s) to a protonation on the NH2 site of the bacterial vitamin p-aminobenzoic acid and forms p-carboxyphenylammonium (or p-carboxyanilinium) cation. The protonation on the cation (NH3+) is well confirmed by the comparison of the C—N bond distance (Tables 1, 3 and 5) with the literature value of 1.38 Å for the unprotonated NH2 group (Lai & Marsh, 1967).
In compounds (I) and (II), the head (NH3+) and tail (COOH) of the cations are connected through glide-related hydrogen bonds, N4—H4A···O1A(−x + 2, y + 1/2, −z + 3/2) and N4—H4A···O1A(x − 1, −y + 1/2, z − 1/2), respectively, leading to a C(8) hydrogen-bonding graph-set motif. In (I), these motifs form a layered structure stacked parallel to the ab plane with an inter-layer distance of 6.695 Å. In (II), aggregation of these C(8) motifs leads to a sheet-like structure on the (102) parallel planes of the unit cell leading to strong X-ray reflections for the corresponding planes. These sheets are stacked with an inter-layer distance of 7.009 Å. From the Cambridge Structural Database, it is observed that a head-to-tail hydrogen-bonding association and carboxylic group dimerization are characterestic features found in most PABA complexes (Allen, 2002). The former feature is observed in (I) and (II) and the later in (III). In (III), the carboxyl groups of the cations are dimerized through O1B—H1B···O2A and O2B···H2B···O1A hydrogen bonds, leading to a graph-set motif of R22(8). Schematic diagrams of different hydrogen-bond motifs are shown in Fig. 4.
In all three structures, twisting of the carboxyl plane from the aromatic ring is observed. The angle of twisting is 9.55 (9)° in (I), 6.25 (3)° in (II), and 5.95 (3) and 3.41 (2)° for the cations in (III). In structure (II), atom O1A deviates by 0.151 (4) Å, which is more than the deviation of O1B (the hydroxy atom), whereas in structure (I) and both the residues of structure (III), atom O1B deviates [0.277 (7) Å in (I), and 0.215 (7) and 0.121 (6) Å in (III)] more than O1A. This twisting of the carboxylic acid group is due to the hydrogen-bonding association and the packing specificity of the crystal. In all three structures, the cations are linked by the anions through N—H···O bonds to form an infinite chain running along the c axis with the linear C22(6) graph-set motif [N4—H4C···O1/N4—H4B···O2 (x, y, z + 1) in (I), N4—H4B···O2(x − 1, −y + 1/2, z+1/2)/N4—H4C···O3(x − 1, −y + 1/2, z − 1/2) in (II), and N14—H14B···O2(x, y, z − 1)/N14—H14C···O1 and N24—H24A···O4(1 + x, y, 1 + z)/N24—H24C···O3(1 + x, y, z) in both the cations of (III).
The packing diagram of (I) in Fig. 5 shows hydrophobic layers formed by the stacking of aromatic rings at x = 0. Also in (I), a hydrogen-bonding graph-set motif of R64(16) exists as a result of glide-related direct interactions and interactions tinvolving the anions and the amine and carboxylic groups. These closed rings of size 2.850 × 8.681 Å (including contact radii) extend along the c axis. This closed-ring R64(16) structure results in a pseudo-inversion centre at (1/2, 1/2, 0), which is extended as a pseudo-inversion axis through (1/2, 1/2, 1/2) (Fig. 6). Such pseudo-symmetries are also observed in the structure of PABA (Lai & Marsh, 1967). Fig. 7 represents the structural overlay of the cation in structure (I) (minor occupancy of OH group is omitted) with both the residues of structure (III) by fitting together the atoms of the aromatic ring. Slight deviation of the amino and carboxyl groups of the cations in (I) is observed.
In (II), perchlorate anions are sandwiched between hydrophobic layers of cations at y = 1/4 and 3/4, thus forming alternating hydrophilic and hydrophobic columns in the bc plane of the unit cell (Fig. 8). Primary C22(6) graph-set motifs connect two adjacent sheets of the p-carboxyphenylammonium ion formed by the above-mentioned C(8) primary graph-set motif. Combination of these two primary motifs leads to a new secondary graph-set motif R66(28) (Fig. 9). The water molecules interact with the anions through weak hydrogen bonds, connecting two inversely related perchlorate anions to form a closed hydrogen-bonding dimer or ring, leading to a graph-set motif of R44(12) (Fig. 10). These motifs are connected linearly along the c axis by another weak interaction O1W···O4(−x + 1, −y + 1, −z + 1) [3.162 (4) Å]. Three-centered hydrogen bonds, formed by the amino group with the anion, relate two inversely related cations through weak hydrogen bonds [N4—H4C···O1(x − 1, −y + 1/2, z + 1/2) and N4—H4C···O3(1 − x, 1/2 + y, 3/2 − z)] making a graph-set motif of R24(10).
In (III), the sulfate anions are stacked nearly at x = 0 forming hydrophilic columns. Alternatively, hydrophobic layers are observed nearly at x = 1/2 and 3/4 owing to the stacking of aromatic rings (Fig. 11). The anions are connected to the cationic dimers through extensive hydrogen bonding between the O atoms of the anion and the amino group. The cationic dimers connected by the above-mentioned C22(6) hydrogen-bond motifs form a sheet-like structure parallel to the ac plane (Fig. 12). The planes of the cationic dimers make a dihedral angle of 20.87° with the ac plane of the unit cell.