As models for highly concentrated biological and other colloidal systems, computer simulations using Monte Carlo (MC) methods are used to study two bidisperse systems of spheres, one system consisting of mixtures of hard spheres and the other consisting of mixtures of hard spheres and spheres having an additional repulsive potential. First, the nonideal parts of the chemical potentials 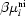 (i = 1, 2) are obtained by using Widom's test particle method. From the chemical potentials the osmotic compression factor Z is obtained by Gibbs–Duhem integration of
(i = 1, 2) are obtained by using Widom's test particle method. From the chemical potentials the osmotic compression factor Z is obtained by Gibbs–Duhem integration of 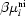 . For the hard-particle model, comparison is also made with Z values calculated from the pair correlation functions pij at contact, and with Z values obtained using an analytical expression based on Percus–Yevick theory. Next, the normalized isothermal compressibility ρkTχT is obtained from
. For the hard-particle model, comparison is also made with Z values calculated from the pair correlation functions pij at contact, and with Z values obtained using an analytical expression based on Percus–Yevick theory. Next, the normalized isothermal compressibility ρkTχT is obtained from 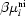 , via the Gibbs–Duhem values of Z. During the MC simulations the partial structure factors Sij are also calculated by Debye double summation and by averaging over the cubic MC cell. The latter parameters are used to calculate the corresponding small-angle X-ray and neutron intensities I(Q), which are compared with intensities obtained by Percus–Yevick theory. Comparison is also made between the structural values of
, via the Gibbs–Duhem values of Z. During the MC simulations the partial structure factors Sij are also calculated by Debye double summation and by averaging over the cubic MC cell. The latter parameters are used to calculate the corresponding small-angle X-ray and neutron intensities I(Q), which are compared with intensities obtained by Percus–Yevick theory. Comparison is also made between the structural values of 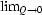 I(Q) obtained by Debye double summation and the thermodynamic ρkTχT values obtained from
I(Q) obtained by Debye double summation and the thermodynamic ρkTχT values obtained from 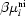 . As already seen, e.g. from measurements of osmotic pressure, the MC simulations show that, for a typical biological system, activity factors can deviate from the value one by several powers of ten, already at fairly low concentrations.
. As already seen, e.g. from measurements of osmotic pressure, the MC simulations show that, for a typical biological system, activity factors can deviate from the value one by several powers of ten, already at fairly low concentrations.

 journal menu
journal menu











