
We obtained two conformational polymorphs of 2,5-dichloro-3,6-bis(dibenzylamino)-
p-hydroquinone, C
34H
30Cl
2N
2O
2. Both polymorphs have an inversion centre at the centre of the hydroquinone ring (
Z′ =
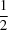
), and there are no significant differences between their bond lengths and angles. The most significant structural difference in the molecular conformations was found in the rotation of the phenyl rings of the two crystallographically independent benzyl groups. The crystal structures of the polymorphs were distinguishable with respect to the arrangement of the hydroquinone rings and the packing motif of the phenyl rings that form part of the benzyl groups. The phenyl groups of one polymorph are arranged in a face-to-edge motif between adjacent molecules, with intermolecular C—H

π interactions, whereas the phenyl rings in the other polymorph form a lamellar stacking pattern with no significant intermolecular interactions. We suggest that this partial conformational difference in the molecular structures leads to the significant structural differences observed in their molecular arrangements.
Supporting information
CCDC references: 1826861; 1826860
For both structures, data collection: RAPID-AUTO (Rigaku, 2000); cell refinement: RAPID-AUTO (Rigaku, 2000); data reduction: RAPID-AUTO (Rigaku, 2000); program(s) used to solve structure: SHELXT (Sheldrick, 2015a); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2015b); molecular graphics: CrystalStructure (Rigaku, 2017) and Mercury (Macrae et al., 2008); software used to prepare material for publication: CrystalStructure (Rigaku, 2017).
2,5-Bis(dibenzylamino)-3,6-dichlorobenzene-1,4-diol (20130730_colorless_block)
top
Crystal data top
| C34H30Cl2N2O2 | F(000) = 596.00 |
| Mr = 569.53 | Dx = 1.319 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54187 Å |
| a = 9.67404 (18) Å | Cell parameters from 10981 reflections |
| b = 10.9707 (2) Å | θ = 3.3–68.2° |
| c = 13.5552 (3) Å | µ = 2.30 mm−1 |
| β = 94.7380 (7)° | T = 296 K |
| V = 1433.72 (5) Å3 | Block, colorless |
| Z = 2 | 0.40 × 0.33 × 0.23 mm |
Data collection top
Rigaku R-AXIS RAPID
diffractometer | 2101 reflections with F2 > 2.0σ(F2) |
| Detector resolution: 10.000 pixels mm-1 | Rint = 0.070 |
| ω scans | θmax = 66.5°, θmin = 5.2° |
Absorption correction: multi-scan
(ABSCOR; Rigaku, 1995) | h = −11→11 |
| Tmin = 0.492, Tmax = 0.589 | k = −13→12 |
| 12895 measured reflections | l = −16→16 |
| 2527 independent reflections | |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.042 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.125 | H-atom parameters constrained |
| S = 1.10 | w = 1/[σ2(Fo2) + (0.0573P)2 + 0.3915P]
where P = (Fo2 + 2Fc2)/3 |
| 2527 reflections | (Δ/σ)max < 0.001 |
| 181 parameters | Δρmax = 0.20 e Å−3 |
| 0 restraints | Δρmin = −0.32 e Å−3 |
| Primary atom site location: structure-invariant direct methods | |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement was performed using all reflections. The weighted
R-factor (wR) and goodness of fit (S) are based on F2.
R-factor (gt) are based on F. The threshold expression of
F2 > 2.0 sigma(F2) is used only for calculating R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cl1 | 0.23763 (5) | 0.60878 (5) | 0.58672 (5) | 0.0671 (2) | |
| O1 | 0.50864 (15) | 0.25956 (12) | 0.44492 (11) | 0.0578 (4) | |
| H1 | 0.4330 | 0.2280 | 0.4510 | 0.069* | |
| N1 | 0.27948 (15) | 0.33329 (14) | 0.52062 (11) | 0.0471 (4) | |
| C1 | 0.38157 (19) | 0.54615 (17) | 0.53814 (14) | 0.0472 (5) | |
| C2 | 0.50464 (19) | 0.37915 (16) | 0.47231 (14) | 0.0456 (4) | |
| C3 | 0.38501 (18) | 0.42424 (17) | 0.51024 (14) | 0.0443 (4) | |
| C4 | 0.2697 (2) | 0.2951 (2) | 0.62473 (15) | 0.0563 (5) | |
| H4A | 0.2169 | 0.3553 | 0.6582 | 0.068* | |
| H4B | 0.3621 | 0.2916 | 0.6584 | 0.068* | |
| C5 | 0.2012 (2) | 0.17238 (19) | 0.63111 (14) | 0.0511 (5) | |
| C6 | 0.2621 (3) | 0.0708 (2) | 0.5945 (2) | 0.0787 (8) | |
| H6 | 0.3439 | 0.0794 | 0.5636 | 0.094* | |
| C7 | 0.2052 (4) | −0.0436 (3) | 0.6023 (2) | 0.0935 (9) | |
| H7 | 0.2487 | −0.1109 | 0.5769 | 0.112* | |
| C8 | 0.0857 (3) | −0.0581 (3) | 0.6468 (2) | 0.0825 (8) | |
| H8 | 0.0466 | −0.1351 | 0.6515 | 0.099* | |
| C9 | 0.0231 (3) | 0.0415 (3) | 0.6847 (2) | 0.0894 (9) | |
| H9 | −0.0586 | 0.0320 | 0.7155 | 0.107* | |
| C10 | 0.0815 (3) | 0.1569 (2) | 0.6773 (2) | 0.0747 (7) | |
| H10 | 0.0389 | 0.2239 | 0.7038 | 0.090* | |
| C11 | 0.14278 (19) | 0.3557 (2) | 0.46778 (15) | 0.0534 (5) | |
| H11A | 0.1028 | 0.4281 | 0.4950 | 0.064* | |
| H11B | 0.0823 | 0.2876 | 0.4794 | 0.064* | |
| C12 | 0.14744 (19) | 0.37228 (19) | 0.35847 (15) | 0.0513 (5) | |
| C13 | 0.1040 (3) | 0.4804 (2) | 0.31389 (18) | 0.0696 (6) | |
| H13 | 0.0771 | 0.5446 | 0.3528 | 0.084* | |
| C14 | 0.1000 (3) | 0.4945 (3) | 0.2136 (2) | 0.0937 (9) | |
| H14 | 0.0698 | 0.5678 | 0.1848 | 0.112* | |
| C15 | 0.1400 (3) | 0.4016 (4) | 0.1557 (2) | 0.0994 (11) | |
| H15 | 0.1376 | 0.4119 | 0.0875 | 0.119* | |
| C16 | 0.1842 (3) | 0.2922 (3) | 0.1976 (2) | 0.0884 (9) | |
| H16 | 0.2115 | 0.2288 | 0.1580 | 0.106* | |
| C17 | 0.1872 (2) | 0.2781 (2) | 0.29918 (18) | 0.0653 (6) | |
| H17 | 0.2164 | 0.2045 | 0.3279 | 0.078* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl1 | 0.0469 (3) | 0.0551 (4) | 0.1018 (5) | 0.0003 (2) | 0.0209 (3) | −0.0140 (3) |
| O1 | 0.0512 (8) | 0.0380 (7) | 0.0852 (10) | −0.0062 (6) | 0.0105 (7) | −0.0088 (7) |
| N1 | 0.0411 (8) | 0.0449 (9) | 0.0552 (9) | −0.0097 (7) | 0.0031 (7) | 0.0023 (7) |
| C1 | 0.0386 (9) | 0.0423 (11) | 0.0608 (11) | 0.0005 (8) | 0.0050 (8) | −0.0005 (8) |
| C2 | 0.0429 (10) | 0.0369 (10) | 0.0566 (11) | −0.0014 (7) | 0.0023 (8) | −0.0035 (8) |
| C3 | 0.0383 (9) | 0.0405 (10) | 0.0539 (10) | −0.0044 (7) | 0.0020 (8) | 0.0012 (8) |
| C4 | 0.0623 (13) | 0.0525 (12) | 0.0539 (11) | −0.0106 (10) | 0.0034 (9) | 0.0018 (9) |
| C5 | 0.0507 (11) | 0.0511 (12) | 0.0513 (11) | −0.0068 (9) | 0.0040 (8) | 0.0062 (9) |
| C6 | 0.0799 (17) | 0.0577 (15) | 0.104 (2) | −0.0041 (12) | 0.0393 (15) | 0.0087 (13) |
| C7 | 0.121 (2) | 0.0533 (16) | 0.112 (2) | −0.0106 (15) | 0.0447 (19) | 0.0034 (14) |
| C8 | 0.0879 (19) | 0.0638 (17) | 0.0942 (19) | −0.0256 (15) | −0.0020 (15) | 0.0164 (14) |
| C9 | 0.0590 (15) | 0.094 (2) | 0.117 (2) | −0.0174 (14) | 0.0193 (14) | 0.0338 (18) |
| C10 | 0.0664 (15) | 0.0677 (16) | 0.0934 (18) | 0.0009 (12) | 0.0275 (13) | 0.0113 (13) |
| C11 | 0.0388 (10) | 0.0586 (12) | 0.0628 (12) | −0.0069 (9) | 0.0028 (9) | 0.0030 (9) |
| C12 | 0.0378 (10) | 0.0537 (12) | 0.0616 (12) | −0.0105 (8) | −0.0010 (8) | 0.0042 (9) |
| C13 | 0.0696 (15) | 0.0632 (15) | 0.0751 (15) | −0.0053 (12) | 0.0002 (12) | 0.0099 (12) |
| C14 | 0.097 (2) | 0.095 (2) | 0.087 (2) | −0.0101 (18) | −0.0032 (16) | 0.0305 (18) |
| C15 | 0.086 (2) | 0.147 (3) | 0.0642 (17) | −0.020 (2) | 0.0021 (15) | 0.0167 (19) |
| C16 | 0.0723 (17) | 0.118 (3) | 0.0746 (17) | −0.0085 (17) | 0.0060 (13) | −0.0263 (17) |
| C17 | 0.0573 (13) | 0.0657 (15) | 0.0722 (15) | −0.0038 (11) | 0.0006 (11) | −0.0074 (12) |
Geometric parameters (Å, º) top
| Cl1—C1 | 1.7309 (19) | C8—C9 | 1.369 (4) |
| O1—C2 | 1.365 (2) | C8—H8 | 0.9300 |
| O1—H1 | 0.8200 | C9—C10 | 1.393 (4) |
| N1—C3 | 1.443 (2) | C9—H9 | 0.9300 |
| N1—C11 | 1.472 (2) | C10—H10 | 0.9300 |
| N1—C4 | 1.482 (2) | C11—C12 | 1.497 (3) |
| C1—C2i | 1.389 (3) | C11—H11A | 0.9700 |
| C1—C3 | 1.391 (3) | C11—H11B | 0.9700 |
| C2—C1i | 1.389 (3) | C12—C13 | 1.381 (3) |
| C2—C3 | 1.395 (3) | C12—C17 | 1.383 (3) |
| C4—C5 | 1.507 (3) | C13—C14 | 1.365 (4) |
| C4—H4A | 0.9700 | C13—H13 | 0.9300 |
| C4—H4B | 0.9700 | C14—C15 | 1.363 (5) |
| C5—C10 | 1.372 (3) | C14—H14 | 0.9300 |
| C5—C6 | 1.372 (3) | C15—C16 | 1.380 (5) |
| C6—C7 | 1.378 (4) | C15—H15 | 0.9300 |
| C6—H6 | 0.9300 | C16—C17 | 1.384 (4) |
| C7—C8 | 1.356 (4) | C16—H16 | 0.9300 |
| C7—H7 | 0.9300 | C17—H17 | 0.9300 |
| | | |
| C2—O1—H1 | 109.5 | C8—C9—C10 | 120.2 (3) |
| C3—N1—C11 | 116.86 (15) | C8—C9—H9 | 119.9 |
| C3—N1—C4 | 112.92 (14) | C10—C9—H9 | 119.9 |
| C11—N1—C4 | 112.65 (15) | C5—C10—C9 | 120.5 (3) |
| C2i—C1—C3 | 120.17 (17) | C5—C10—H10 | 119.7 |
| C2i—C1—Cl1 | 118.29 (15) | C9—C10—H10 | 119.7 |
| C3—C1—Cl1 | 121.55 (14) | N1—C11—C12 | 113.74 (16) |
| O1—C2—C1i | 119.93 (17) | N1—C11—H11A | 108.8 |
| O1—C2—C3 | 118.95 (17) | C12—C11—H11A | 108.8 |
| C1i—C2—C3 | 121.12 (17) | N1—C11—H11B | 108.8 |
| C1—C3—C2 | 118.71 (17) | C12—C11—H11B | 108.8 |
| C1—C3—N1 | 127.26 (17) | H11A—C11—H11B | 107.7 |
| C2—C3—N1 | 113.95 (16) | C13—C12—C17 | 118.5 (2) |
| N1—C4—C5 | 111.72 (16) | C13—C12—C11 | 120.3 (2) |
| N1—C4—H4A | 109.3 | C17—C12—C11 | 121.1 (2) |
| C5—C4—H4A | 109.3 | C14—C13—C12 | 121.1 (3) |
| N1—C4—H4B | 109.3 | C14—C13—H13 | 119.5 |
| C5—C4—H4B | 109.3 | C12—C13—H13 | 119.5 |
| H4A—C4—H4B | 107.9 | C15—C14—C13 | 120.2 (3) |
| C10—C5—C6 | 117.9 (2) | C15—C14—H14 | 119.9 |
| C10—C5—C4 | 122.0 (2) | C13—C14—H14 | 119.9 |
| C6—C5—C4 | 120.0 (2) | C14—C15—C16 | 120.4 (3) |
| C5—C6—C7 | 121.7 (2) | C14—C15—H15 | 119.8 |
| C5—C6—H6 | 119.2 | C16—C15—H15 | 119.8 |
| C7—C6—H6 | 119.2 | C15—C16—C17 | 119.2 (3) |
| C8—C7—C6 | 120.1 (3) | C15—C16—H16 | 120.4 |
| C8—C7—H7 | 119.9 | C17—C16—H16 | 120.4 |
| C6—C7—H7 | 119.9 | C12—C17—C16 | 120.7 (3) |
| C7—C8—C9 | 119.5 (3) | C12—C17—H17 | 119.7 |
| C7—C8—H8 | 120.3 | C16—C17—H17 | 119.7 |
| C9—C8—H8 | 120.3 | | |
| | | |
| C2i—C1—C3—C2 | 0.3 (3) | C5—C6—C7—C8 | 0.2 (5) |
| Cl1—C1—C3—C2 | −179.49 (14) | C6—C7—C8—C9 | −0.7 (5) |
| C2i—C1—C3—N1 | 176.67 (17) | C7—C8—C9—C10 | 0.2 (5) |
| Cl1—C1—C3—N1 | −3.1 (3) | C6—C5—C10—C9 | −1.1 (4) |
| O1—C2—C3—C1 | 179.93 (17) | C4—C5—C10—C9 | −177.9 (2) |
| C1i—C2—C3—C1 | −0.3 (3) | C8—C9—C10—C5 | 0.7 (4) |
| O1—C2—C3—N1 | 3.1 (3) | C3—N1—C11—C12 | 57.2 (2) |
| C1i—C2—C3—N1 | −177.14 (17) | C4—N1—C11—C12 | −169.59 (17) |
| C11—N1—C3—C1 | 61.2 (3) | N1—C11—C12—C13 | −118.9 (2) |
| C4—N1—C3—C1 | −71.9 (2) | N1—C11—C12—C17 | 64.9 (3) |
| C11—N1—C3—C2 | −122.31 (19) | C17—C12—C13—C14 | 0.1 (3) |
| C4—N1—C3—C2 | 104.6 (2) | C11—C12—C13—C14 | −176.1 (2) |
| C3—N1—C4—C5 | −158.83 (17) | C12—C13—C14—C15 | −0.5 (4) |
| C11—N1—C4—C5 | 66.1 (2) | C13—C14—C15—C16 | 0.4 (5) |
| N1—C4—C5—C10 | −120.2 (2) | C14—C15—C16—C17 | 0.0 (5) |
| N1—C4—C5—C6 | 63.1 (3) | C13—C12—C17—C16 | 0.3 (3) |
| C10—C5—C6—C7 | 0.7 (4) | C11—C12—C17—C16 | 176.5 (2) |
| C4—C5—C6—C7 | 177.6 (3) | C15—C16—C17—C12 | −0.3 (4) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.82 | 2.16 | 2.645 (2) | 118 |
2,5-Bis(dibenzylamino)-3,6-dichlorobenzene-1,4-diol (20130722_palered_block)
top
Crystal data top
| C34H30Cl2N2O2 | Dx = 1.366 Mg m−3 |
| Mr = 569.53 | Cu Kα radiation, λ = 1.54187 Å |
| Orthorhombic, Pbca | Cell parameters from 17342 reflections |
| a = 13.2054 (2) Å | θ = 3.4–68.2° |
| b = 12.4456 (2) Å | µ = 2.39 mm−1 |
| c = 16.8489 (3) Å | T = 296 K |
| V = 2769.09 (9) Å3 | Block, pale red |
| Z = 4 | 0.40 × 0.30 × 0.28 mm |
| F(000) = 1192.00 | |
Data collection top
Rigaku R-AXIS RAPID
diffractometer | 1976 reflections with F2 > 2.0σ(F2) |
| Detector resolution: 10.000 pixels mm-1 | Rint = 0.039 |
| ω scans | θmax = 66.5°, θmin = 5.3° |
Absorption correction: multi-scan
(ABSCOR; Rigaku, 1995) | h = −15→15 |
| Tmin = 0.413, Tmax = 0.507 | k = −14→14 |
| 23298 measured reflections | l = −20→20 |
| 2442 independent reflections | |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.115 | H-atom parameters constrained |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0633P)2 + 0.3919P]
where P = (Fo2 + 2Fc2)/3 |
| 2442 reflections | (Δ/σ)max < 0.001 |
| 181 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
| Primary atom site location: structure-invariant direct methods | |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refinement was performed using all reflections. The weighted
R-factor (wR) and goodness of fit (S) are based on F2.
R-factor (gt) are based on F. The threshold expression of
F2 > 2.0 sigma(F2) is used only for calculating R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| Cl1 | 0.66299 (4) | 0.47433 (4) | 0.63253 (3) | 0.0591 (2) | |
| O1 | 0.54700 (11) | 0.30198 (9) | 0.56222 (9) | 0.0591 (4) | |
| H1 | 0.5169 | 0.2548 | 0.5378 | 0.071* | |
| N1 | 0.59086 (11) | 0.69385 (10) | 0.55443 (9) | 0.0442 (4) | |
| C1 | 0.57303 (13) | 0.49107 (13) | 0.55909 (11) | 0.0436 (4) | |
| C2 | 0.52341 (13) | 0.40005 (13) | 0.53069 (11) | 0.0426 (4) | |
| C3 | 0.54977 (13) | 0.59272 (13) | 0.52829 (11) | 0.0404 (4) | |
| C4 | 0.55436 (16) | 0.72490 (15) | 0.63403 (12) | 0.0534 (5) | |
| H4A | 0.5935 | 0.6863 | 0.6736 | 0.064* | |
| H4B | 0.4843 | 0.7025 | 0.6394 | 0.064* | |
| C5 | 0.56120 (13) | 0.84400 (14) | 0.65136 (11) | 0.0463 (4) | |
| C6 | 0.56417 (15) | 0.87817 (16) | 0.72920 (12) | 0.0559 (5) | |
| H6 | 0.5655 | 0.8277 | 0.7699 | 0.067* | |
| C7 | 0.56520 (17) | 0.98641 (18) | 0.74745 (14) | 0.0663 (6) | |
| H7 | 0.5671 | 1.0082 | 0.8002 | 0.080* | |
| C8 | 0.56340 (16) | 1.06209 (17) | 0.68797 (15) | 0.0662 (6) | |
| H8 | 0.5633 | 1.1349 | 0.7004 | 0.079* | |
| C9 | 0.56174 (16) | 1.02952 (15) | 0.61040 (15) | 0.0601 (6) | |
| H9 | 0.5618 | 1.0803 | 0.5699 | 0.072* | |
| C10 | 0.56001 (15) | 0.92137 (15) | 0.59207 (13) | 0.0543 (5) | |
| H10 | 0.5580 | 0.9001 | 0.5392 | 0.065* | |
| C11 | 0.70119 (14) | 0.70866 (14) | 0.54321 (12) | 0.0515 (5) | |
| H11A | 0.7369 | 0.6508 | 0.5698 | 0.062* | |
| H11B | 0.7217 | 0.7757 | 0.5677 | 0.062* | |
| C12 | 0.73094 (14) | 0.71014 (13) | 0.45730 (12) | 0.0497 (5) | |
| C13 | 0.71914 (18) | 0.80139 (16) | 0.41111 (15) | 0.0698 (7) | |
| H13 | 0.6907 | 0.8626 | 0.4335 | 0.084* | |
| C14 | 0.7485 (2) | 0.80329 (19) | 0.33315 (16) | 0.0790 (7) | |
| H14 | 0.7399 | 0.8656 | 0.3034 | 0.095* | |
| C15 | 0.79067 (18) | 0.71382 (19) | 0.29857 (14) | 0.0718 (7) | |
| H15 | 0.8106 | 0.7152 | 0.2457 | 0.086* | |
| C16 | 0.80286 (17) | 0.62302 (19) | 0.34285 (15) | 0.0689 (6) | |
| H16 | 0.8316 | 0.5622 | 0.3200 | 0.083* | |
| C17 | 0.77298 (16) | 0.62081 (16) | 0.42091 (14) | 0.0609 (6) | |
| H17 | 0.7812 | 0.5579 | 0.4500 | 0.073* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Cl1 | 0.0653 (4) | 0.0469 (3) | 0.0651 (4) | 0.0030 (2) | −0.0185 (2) | 0.0012 (2) |
| O1 | 0.0731 (9) | 0.0293 (6) | 0.0748 (10) | 0.0011 (6) | −0.0143 (7) | 0.0051 (6) |
| N1 | 0.0484 (9) | 0.0320 (7) | 0.0523 (9) | −0.0043 (6) | 0.0044 (7) | −0.0072 (6) |
| C1 | 0.0475 (10) | 0.0361 (9) | 0.0472 (11) | 0.0015 (7) | −0.0018 (8) | −0.0018 (8) |
| C2 | 0.0497 (10) | 0.0293 (8) | 0.0488 (11) | 0.0029 (7) | 0.0014 (8) | 0.0015 (7) |
| C3 | 0.0448 (9) | 0.0303 (8) | 0.0461 (10) | −0.0006 (7) | 0.0037 (8) | −0.0030 (7) |
| C4 | 0.0672 (13) | 0.0384 (10) | 0.0546 (12) | −0.0027 (9) | 0.0083 (9) | −0.0064 (8) |
| C5 | 0.0478 (10) | 0.0376 (10) | 0.0534 (11) | 0.0008 (8) | 0.0010 (8) | −0.0071 (8) |
| C6 | 0.0621 (12) | 0.0518 (11) | 0.0537 (12) | −0.0002 (9) | 0.0009 (9) | −0.0068 (9) |
| C7 | 0.0749 (14) | 0.0604 (13) | 0.0637 (16) | −0.0002 (11) | 0.0003 (11) | −0.0239 (11) |
| C8 | 0.0678 (14) | 0.0420 (11) | 0.0888 (17) | 0.0004 (10) | −0.0005 (12) | −0.0189 (12) |
| C9 | 0.0671 (14) | 0.0389 (11) | 0.0744 (16) | 0.0021 (9) | −0.0025 (11) | −0.0023 (10) |
| C10 | 0.0635 (13) | 0.0431 (11) | 0.0564 (12) | −0.0002 (9) | −0.0032 (10) | −0.0061 (9) |
| C11 | 0.0492 (11) | 0.0435 (10) | 0.0619 (13) | −0.0062 (8) | 0.0030 (9) | −0.0064 (9) |
| C12 | 0.0455 (11) | 0.0384 (9) | 0.0651 (13) | −0.0057 (7) | 0.0057 (9) | −0.0060 (9) |
| C13 | 0.0837 (16) | 0.0427 (11) | 0.0831 (17) | 0.0026 (10) | 0.0273 (13) | 0.0010 (10) |
| C14 | 0.0929 (18) | 0.0616 (14) | 0.0824 (17) | −0.0039 (12) | 0.0206 (15) | 0.0147 (13) |
| C15 | 0.0717 (15) | 0.0784 (17) | 0.0653 (15) | −0.0159 (12) | 0.0136 (12) | −0.0100 (13) |
| C16 | 0.0688 (14) | 0.0613 (14) | 0.0768 (17) | −0.0043 (11) | 0.0125 (12) | −0.0224 (12) |
| C17 | 0.0637 (13) | 0.0453 (11) | 0.0738 (15) | 0.0015 (9) | 0.0048 (11) | −0.0072 (10) |
Geometric parameters (Å, º) top
| Cl1—C1 | 1.7280 (19) | C8—C9 | 1.368 (3) |
| O1—C2 | 1.367 (2) | C8—H8 | 0.9300 |
| O1—H1 | 0.8200 | C9—C10 | 1.381 (3) |
| N1—C3 | 1.440 (2) | C9—H9 | 0.9300 |
| N1—C4 | 1.477 (2) | C10—H10 | 0.9300 |
| N1—C11 | 1.481 (2) | C11—C12 | 1.500 (3) |
| C1—C2 | 1.393 (2) | C11—H11A | 0.9700 |
| C1—C3 | 1.402 (2) | C11—H11B | 0.9700 |
| C2—C3i | 1.389 (3) | C12—C13 | 1.385 (3) |
| C3—C2i | 1.389 (3) | C12—C17 | 1.386 (3) |
| C4—C5 | 1.514 (2) | C13—C14 | 1.370 (3) |
| C4—H4A | 0.9700 | C13—H13 | 0.9300 |
| C4—H4B | 0.9700 | C14—C15 | 1.374 (3) |
| C5—C6 | 1.379 (3) | C14—H14 | 0.9300 |
| C5—C10 | 1.388 (3) | C15—C16 | 1.364 (3) |
| C6—C7 | 1.382 (3) | C15—H15 | 0.9300 |
| C6—H6 | 0.9300 | C16—C17 | 1.373 (3) |
| C7—C8 | 1.376 (3) | C16—H16 | 0.9300 |
| C7—H7 | 0.9300 | C17—H17 | 0.9300 |
| | | |
| C2—O1—H1 | 109.5 | C8—C9—C10 | 120.2 (2) |
| C3—N1—C4 | 112.56 (14) | C8—C9—H9 | 119.9 |
| C3—N1—C11 | 116.14 (13) | C10—C9—H9 | 119.9 |
| C4—N1—C11 | 113.86 (15) | C9—C10—C5 | 121.00 (19) |
| C2—C1—C3 | 120.24 (17) | C9—C10—H10 | 119.5 |
| C2—C1—Cl1 | 118.11 (13) | C5—C10—H10 | 119.5 |
| C3—C1—Cl1 | 121.65 (13) | N1—C11—C12 | 112.48 (16) |
| O1—C2—C3i | 119.62 (15) | N1—C11—H11A | 109.1 |
| O1—C2—C1 | 119.03 (16) | C12—C11—H11A | 109.1 |
| C3i—C2—C1 | 121.35 (15) | N1—C11—H11B | 109.1 |
| C2i—C3—C1 | 118.41 (15) | C12—C11—H11B | 109.1 |
| C2i—C3—N1 | 115.12 (14) | H11A—C11—H11B | 107.8 |
| C1—C3—N1 | 126.39 (16) | C13—C12—C17 | 117.0 (2) |
| N1—C4—C5 | 114.33 (15) | C13—C12—C11 | 121.51 (17) |
| N1—C4—H4A | 108.7 | C17—C12—C11 | 121.47 (17) |
| C5—C4—H4A | 108.7 | C14—C13—C12 | 121.4 (2) |
| N1—C4—H4B | 108.7 | C14—C13—H13 | 119.3 |
| C5—C4—H4B | 108.7 | C12—C13—H13 | 119.3 |
| H4A—C4—H4B | 107.6 | C13—C14—C15 | 120.5 (2) |
| C6—C5—C10 | 118.10 (18) | C13—C14—H14 | 119.8 |
| C6—C5—C4 | 119.15 (17) | C15—C14—H14 | 119.8 |
| C10—C5—C4 | 122.69 (17) | C16—C15—C14 | 119.2 (2) |
| C5—C6—C7 | 120.8 (2) | C16—C15—H15 | 120.4 |
| C5—C6—H6 | 119.6 | C14—C15—H15 | 120.4 |
| C7—C6—H6 | 119.6 | C15—C16—C17 | 120.4 (2) |
| C8—C7—C6 | 120.3 (2) | C15—C16—H16 | 119.8 |
| C8—C7—H7 | 119.8 | C17—C16—H16 | 119.8 |
| C6—C7—H7 | 119.8 | C16—C17—C12 | 121.5 (2) |
| C9—C8—C7 | 119.6 (2) | C16—C17—H17 | 119.2 |
| C9—C8—H8 | 120.2 | C12—C17—H17 | 119.2 |
| C7—C8—H8 | 120.2 | | |
| | | |
| C3—C1—C2—O1 | −179.49 (16) | C5—C6—C7—C8 | −0.1 (3) |
| Cl1—C1—C2—O1 | 0.5 (2) | C6—C7—C8—C9 | −0.7 (3) |
| C3—C1—C2—C3i | 0.0 (3) | C7—C8—C9—C10 | 1.2 (3) |
| Cl1—C1—C2—C3i | −179.99 (14) | C8—C9—C10—C5 | −0.8 (3) |
| C2—C1—C3—C2i | 0.0 (3) | C6—C5—C10—C9 | 0.0 (3) |
| Cl1—C1—C3—C2i | 179.99 (14) | C4—C5—C10—C9 | 177.01 (19) |
| C2—C1—C3—N1 | 176.65 (16) | C3—N1—C11—C12 | 64.7 (2) |
| Cl1—C1—C3—N1 | −3.4 (3) | C4—N1—C11—C12 | −162.09 (15) |
| C4—N1—C3—C2i | 106.89 (19) | N1—C11—C12—C13 | 80.0 (2) |
| C11—N1—C3—C2i | −119.33 (17) | N1—C11—C12—C17 | −101.1 (2) |
| C4—N1—C3—C1 | −69.8 (2) | C17—C12—C13—C14 | −0.5 (3) |
| C11—N1—C3—C1 | 63.9 (2) | C11—C12—C13—C14 | 178.5 (2) |
| C3—N1—C4—C5 | −157.91 (16) | C12—C13—C14—C15 | 0.1 (4) |
| C11—N1—C4—C5 | 67.2 (2) | C13—C14—C15—C16 | 0.0 (4) |
| N1—C4—C5—C6 | −157.34 (17) | C14—C15—C16—C17 | 0.3 (4) |
| N1—C4—C5—C10 | 25.7 (3) | C15—C16—C17—C12 | −0.8 (3) |
| C10—C5—C6—C7 | 0.4 (3) | C13—C12—C17—C16 | 0.8 (3) |
| C4—C5—C6—C7 | −176.68 (19) | C11—C12—C17—C16 | −178.1 (2) |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1i | 0.82 | 2.20 | 2.680 (2) | 117 |
| Symmetry code: (i) −x+1, −y+1, −z+1. |
Selected bond lengths (Å) for (1a) and (1b) top| Parameter | (1a) | (1b) |
| O1—C2 | 1.365 (2) | 1.367 (2) |
| C2—C3 | 1.395 (3) | 1.389 (3) |
| C1—C3 | 1.391 (3) | 1.402 (2) |
| C1—C2 | 1.389 (3) | 1.393 (2) |
| C3—N1 | 1.443 (2) | 1.440 (2) |
| C1—Cl1 | 1.7309 (19) | 1.7280 (19) |
Parameters (°) for important angles and the dihedral angles in (1a)
and (1b)
HQ, Ph1 are Ph2 are the planes of the central C1–C3/C1i–C3i arene, the
C5–C10 phenyl ring and the C12–C17 phenyl ring, respectively [symmetry code:
(i) -x+1, -y+1, -z+1]. [OK?] top| Parameter | (1a) | (1b) |
| C3—N1—C11 | 116.86 (15) | 116.14 (13) |
| C3—N1—C4 | 112.92 (14) | 112.56 (14) |
| N1—C4—C5 | 111.72 (16) | 114.33 (15) |
| N1—C11—C12 | 113.74 (16) | 112.48 (16) |
| HQ—Ph1 | 11.9 (2) | 43.6 (2) |
| HQ—Ph2 | 74.0 (2) | 65.4 (2) |
| Ph1—Ph2 | 62.12 (13) | 25.63 (10) |
 π interactions, whereas the phenyl rings in the other polymorph form a lamellar stacking pattern with no significant intermolecular interactions. We suggest that this partial conformational difference in the molecular structures leads to the significant structural differences observed in their molecular arrangements.
π interactions, whereas the phenyl rings in the other polymorph form a lamellar stacking pattern with no significant intermolecular interactions. We suggest that this partial conformational difference in the molecular structures leads to the significant structural differences observed in their molecular arrangements.
 journal menu
journal menu











