issue contents
October 2008 issue
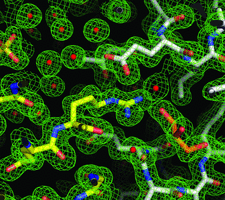
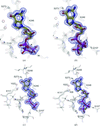
Cover illustration: Crystal-contact engineering of protein crystals, (p. 1020). A systematic study using the diphthine synthase system proved that a single mutation at crystal contact region can improve the diffraction quality of protein crystals. This figure shows an electron-density map around the mutation site of the K49R mutant which provided the best crystals at 1.5 Å resolution as compared with 2.1 Å of wild-type crystals.
research papers
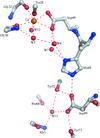
X-ray crystal structures have been obtained and molecular-dynamics simulations have been carried out for three active-site mutants (H48N, D49N and D49K) of bovine pancreatic phospholipase A2. The active site is disturbed in the Asp49 mutant structures.
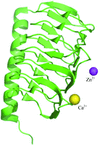
Three crystal structures of the γ-carbonic anhydrase enzyme from P. horikoshii were solved in space group F4132. The enzyme forms a β-helix and each trimer is stacked in a tetrahedral manner in the crystal.
The influence of mutations in crystal-packing regions on X-ray diffraction quality was systematically examined.
PDB references: diphthine synthase, 1wng, r1wngsf; K26R, 2dsg, r2dsgsf; K26Y, 2dsh, r2dshsf; K49R, 2z6r, r2z6rsf; E54H, 2dxv, r2dxvsf; E54K, 2dxw, r2dxwsf; E54R, 2e7r, r2e7rsf; L65A, 2dv5, r2dv5sf; L65Q, 2dv4, r2dv4sf; N69K, 2ehc, r2ehcsf; D79E, 2e07, r2e07sf; E140K, 2e08, r2e08sf; E140N, 2egb, r2egbsf; E140R, 2e16, r2e16sf; N142E, 2dxx, r2dxxsf; T146R, 2ehl, r2ehlsf; E171K, 2egl, r2eglsf; E171R, 2dsi, r2dsisf; R173A, 2e17, r2e17sf; R173N, 2e15, r2e15sf; K187R, 2dv7, r2dv7sf; L261M, 2egs, r2egssf
The structures of pseudechetoxin and pseudecin suggest that both proteins bind to cyclic nucleotide-gated ion channels in a manner in which the concave surface occludes the pore entrance.
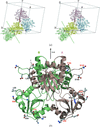
Crystal structures of the Ffh NG GTPase domain at < 1.24 Å resolution reveal multiple overlapping nucleotide binding modes.
The influence of surfaces exposing ionizable groups on the crystallization of model proteins has been investigated.
The crystal structure of an isolated unglycosylated antibody CH2 domain has been determined at 1.7 Å resolution.
PDB reference: antibody CH2 domain, 3dj9, r3dj9sf
The crystal structure of PH1421, a phosphatase belonging to the haloacid dehalogenase superfamily, has been determined at 1.6 Å resolution. The structure reveals a dimeric state which may be important for the catalytic function of the enzyme.
PDB reference: PH1421, 1wr8, r1wr8sf
Peaks in the Patterson maps help in finding molecular positions in the unit cell and assist in molecular-replacement solution in difficult cases for molecules with helical pseudosymmetry.


 journal menu
journal menu