The title compound, C
10H
6O
8.2C
2H
6OS, one of the few known examples of the cocrystallization of pyromellitic acid with common organic solvents, has been previously analysed by single-crystal X-ray diffraction at 296 K [Jin, Pan, Shen, Li & Hu (2003).
Acta Cryst. C
59, o205–o206]. We present here a redetermination at low temperature (150 K). The centrosymmetric compound crystallizes in space group
P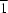
and exhibits co-operative strong (acid)O—H

O(solvent) and weaker (solvent)C—H

O(acid) hydrogen-bonding interactions, giving rise to a one-dimensional ribbon structure along the
bc diagonal of the unit cell.
Supporting information
CCDC reference: 221712
Key indicators
- Single-crystal X-ray study
- T = 150 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.002 Å
(C-C) = 0.002 Å
- R factor = 0.028
- wR factor = 0.075
- Data-to-parameter ratio = 16.8
checkCIF results
No syntax errors found
ADDSYM reports no extra symmetry
 Alert Level B:
Alert Level B:
REFLT_03
From the CIF: _diffrn_reflns_theta_max 29.05
From the CIF: _reflns_number_total 2112
TEST2: Reflns within _diffrn_reflns_theta_max
Count of symmetry unique reflns 2425
Completeness (_total/calc) 87.09%
Alert B: < 90% complete (theta max?)
0 Alert Level A = Potentially serious problem
1 Alert Level B = Potential problem
0 Alert Level C = Please check
Data collection: SMART (Siemens, 1994); cell refinement: SAINT (Siemens, 1994); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Bruker, 1997); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and local programs.
Crystal data top
| C10H6O8·2C2H6OS | Z = 1 |
| Mr = 410.40 | F(000) = 214 |
| Triclinic, P1 | Dx = 1.495 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.3283 (6) Å | Cell parameters from 3039 reflections |
| b = 7.3331 (6) Å | θ = 2.3–29.0° |
| c = 8.9456 (8) Å | µ = 0.34 mm−1 |
| α = 80.510 (2)° | T = 150 K |
| β = 81.124 (2)° | Block, colourless |
| γ = 75.474 (2)° | 0.47 × 0.23 × 0.21 mm |
| V = 455.77 (7) Å3 | |
Data collection top
Bruker SMART 1000 CCD
diffractometer | 2112 independent reflections |
| Radiation source: sealed tube | 1918 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.010 |
| ω rotation with narrow frames scans | θmax = 29.1°, θmin = 2.3° |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2001) | h = −9→9 |
| Tmin = 0.849, Tmax = 0.931 | k = −9→9 |
| 4046 measured reflections | l = −11→11 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: All non-H atoms found by direct methods |
| R[F2 > 2σ(F2)] = 0.028 | Hydrogen site location: Geom except OH coords freely refined |
| wR(F2) = 0.075 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.07 | w = 1/[σ2(Fo2) + (0.0387P)2 + 0.1844P]
where P = (Fo2 + 2Fc2)/3 |
| 2112 reflections | (Δ/σ)max = 0.001 |
| 126 parameters | Δρmax = 0.38 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.56940 (17) | 0.94904 (17) | 0.14218 (13) | 0.0162 (2) | |
| C2 | 0.58863 (17) | 1.11937 (17) | 0.05261 (14) | 0.0168 (2) | |
| H2 | 0.6497 | 1.2009 | 0.0885 | 0.020* | |
| C3 | 0.51965 (17) | 1.17155 (17) | −0.08860 (14) | 0.0160 (2) | |
| C4 | 0.64701 (18) | 0.90580 (17) | 0.29329 (14) | 0.0178 (2) | |
| O1 | 0.54595 (14) | 0.81614 (14) | 0.40204 (10) | 0.0229 (2) | |
| H1 | 0.604 (3) | 0.793 (3) | 0.484 (2) | 0.034* | |
| O2 | 0.78731 (15) | 0.95498 (16) | 0.31123 (11) | 0.0295 (2) | |
| C5 | 0.52586 (18) | 1.36689 (17) | −0.17080 (14) | 0.0174 (2) | |
| O3 | 0.69967 (14) | 1.39404 (14) | −0.19728 (12) | 0.0268 (2) | |
| H3 | 0.704 (3) | 1.496 (3) | −0.238 (2) | 0.040* | |
| O4 | 0.38433 (14) | 1.48471 (14) | −0.20067 (13) | 0.0290 (2) | |
| S1 | 0.88455 (4) | 0.82189 (4) | 0.65791 (4) | 0.01906 (10) | |
| O5 | 0.72689 (13) | 0.72202 (13) | 0.64578 (10) | 0.0217 (2) | |
| C6 | 1.09286 (19) | 0.6984 (2) | 0.55329 (16) | 0.0257 (3) | |
| H6A | 1.1141 | 0.5620 | 0.5895 | 0.039* | |
| H6B | 1.2022 | 0.7447 | 0.5683 | 0.039* | |
| H6C | 1.0769 | 0.7206 | 0.4445 | 0.039* | |
| C7 | 0.9472 (2) | 0.7387 (2) | 0.84614 (16) | 0.0291 (3) | |
| H7A | 0.8374 | 0.7794 | 0.9206 | 0.044* | |
| H7B | 1.0522 | 0.7912 | 0.8617 | 0.044* | |
| H7C | 0.9863 | 0.5996 | 0.8595 | 0.044* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0158 (5) | 0.0178 (6) | 0.0150 (5) | −0.0041 (4) | −0.0023 (4) | −0.0016 (4) |
| C2 | 0.0177 (6) | 0.0165 (6) | 0.0173 (6) | −0.0055 (4) | −0.0032 (4) | −0.0021 (4) |
| C3 | 0.0160 (5) | 0.0158 (5) | 0.0164 (5) | −0.0046 (4) | −0.0017 (4) | −0.0011 (4) |
| C4 | 0.0206 (6) | 0.0166 (6) | 0.0170 (6) | −0.0052 (4) | −0.0047 (4) | −0.0007 (4) |
| O1 | 0.0253 (5) | 0.0295 (5) | 0.0160 (4) | −0.0125 (4) | −0.0064 (4) | 0.0043 (4) |
| O2 | 0.0303 (5) | 0.0405 (6) | 0.0235 (5) | −0.0204 (5) | −0.0117 (4) | 0.0066 (4) |
| C5 | 0.0220 (6) | 0.0173 (6) | 0.0143 (5) | −0.0066 (5) | −0.0037 (4) | −0.0013 (4) |
| O3 | 0.0230 (5) | 0.0219 (5) | 0.0344 (6) | −0.0107 (4) | −0.0051 (4) | 0.0101 (4) |
| O4 | 0.0244 (5) | 0.0184 (5) | 0.0426 (6) | −0.0036 (4) | −0.0098 (4) | 0.0036 (4) |
| S1 | 0.01852 (16) | 0.01855 (16) | 0.02112 (17) | −0.00685 (11) | −0.00345 (11) | −0.00049 (11) |
| O5 | 0.0227 (5) | 0.0241 (5) | 0.0209 (4) | −0.0126 (4) | −0.0072 (4) | 0.0048 (4) |
| C6 | 0.0228 (7) | 0.0299 (7) | 0.0235 (7) | −0.0054 (5) | 0.0004 (5) | −0.0049 (5) |
| C7 | 0.0227 (7) | 0.0480 (9) | 0.0187 (6) | −0.0111 (6) | −0.0037 (5) | −0.0040 (6) |
Geometric parameters (Å, º) top
| C1—C2 | 1.3947 (17) | C5—O3 | 1.3178 (15) |
| C1—C3i | 1.4013 (16) | O3—H3 | 0.78 (2) |
| C1—C4 | 1.5022 (16) | S1—O5 | 1.5403 (9) |
| C2—C3 | 1.3922 (17) | S1—C6 | 1.7808 (14) |
| C2—H2 | 0.9500 | S1—C7 | 1.7825 (14) |
| C3—C5 | 1.5066 (16) | C6—H6A | 0.9800 |
| C4—O2 | 1.2120 (15) | C6—H6B | 0.9800 |
| C4—O1 | 1.3184 (15) | C6—H6C | 0.9800 |
| O1—H1 | 0.87 (2) | C7—H7A | 0.9800 |
| O2—S1 | 3.2270 (10) | C7—H7B | 0.9800 |
| C5—O4 | 1.2056 (16) | C7—H7C | 0.9800 |
| | | |
| C2—C1—C3i | 119.41 (11) | C4—O1—H1 | 107.3 (12) |
| C2—C1—C4 | 117.05 (11) | O4—C5—O3 | 125.33 (12) |
| C3i—C1—C4 | 123.54 (11) | O4—C5—C3 | 122.35 (11) |
| C3—C2—C1 | 120.94 (11) | O3—C5—C3 | 112.22 (10) |
| C3—C2—H2 | 119.5 | C5—O3—H3 | 112.9 (15) |
| C1—C2—H2 | 119.5 | H6A—C6—H6B | 109.5 |
| C2—C3—C1i | 119.64 (11) | H6A—C6—H6C | 109.5 |
| C2—C3—C5 | 117.87 (11) | H6B—C6—H6C | 109.5 |
| C1i—C3—C5 | 122.17 (11) | H7A—C7—H7B | 109.5 |
| O2—C4—O1 | 124.39 (11) | H7A—C7—H7C | 109.5 |
| O2—C4—C1 | 121.73 (11) | H7B—C7—H7C | 109.5 |
| O1—C4—C1 | 113.86 (10) | | |
| | | |
| C3i—C1—C2—C3 | 0.38 (19) | C3i—C1—C4—O1 | −35.24 (16) |
| C4—C1—C2—C3 | −179.55 (11) | O1—C4—O2—S1 | 2.03 (16) |
| C1—C2—C3—C1i | −0.39 (19) | C1—C4—O2—S1 | −179.96 (9) |
| C1—C2—C3—C5 | 173.29 (11) | C2—C3—C5—O4 | −118.06 (14) |
| C2—C1—C4—O2 | −33.53 (17) | C1i—C3—C5—O4 | 55.45 (18) |
| C3i—C1—C4—O2 | 146.55 (13) | C2—C3—C5—O3 | 58.60 (15) |
| C2—C1—C4—O1 | 144.69 (11) | C1i—C3—C5—O3 | −127.89 (13) |
| Symmetry code: (i) −x+1, −y+2, −z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O5 | 0.87 (2) | 1.76 (2) | 2.6317 (13) | 173.9 (18) |
| O3—H3···O5ii | 0.78 (2) | 1.84 (2) | 2.6146 (13) | 173 (2) |
| C6—H6C···O2 | 0.98 | 2.67 | 3.3595 (18) | 127 |
| Symmetry code: (ii) x, y+1, z−1. |
 O(solvent) and weaker (solvent)C—H
O(solvent) and weaker (solvent)C—H O(acid) hydrogen-bonding interactions, giving rise to a one-dimensional ribbon structure along the bc diagonal of the unit cell.
O(acid) hydrogen-bonding interactions, giving rise to a one-dimensional ribbon structure along the bc diagonal of the unit cell.![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.002 Å
(C-C) = 0.002 ÅAlert Level B:

 journal menu
journal menu













