The title compound, C
18H
24N
2O
4, is an aminonaphthoquinone derivative that exhibits a large third-order non-linear optical susceptibility. The centrosymmetric naphthoquinone moieties are connected by bifurcated N—H

O hydrogen bonds to form a ribbon structure along [1
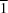
0]. The molecules are stacked along the
c axis, with overlap occurring only at the periphery of the naphthoquinone skeleton.
Supporting information
CCDC reference: 225722
Key indicators
- Single-crystal X-ray study
- T = 93 K
- Mean
![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.003 Å
(C-C) = 0.003 Å
- R factor = 0.047
- wR factor = 0.117
- Data-to-parameter ratio = 12.7
checkCIF/PLATON results
No syntax errors found
 Alert level A
PLAT029_ALERT_3_A _diffrn_measured_fraction_theta_full Low ....... 0.91
Alert level A
PLAT029_ALERT_3_A _diffrn_measured_fraction_theta_full Low ....... 0.91
 Alert level C
REFLT03_ALERT_3_C Reflection count < 95% complete
From the CIF: _diffrn_reflns_theta_max 68.19
From the CIF: _diffrn_reflns_theta_full 0.00
From the CIF: _reflns_number_total 1429
TEST2: Reflns within _diffrn_reflns_theta_max
Count of symmetry unique reflns 1584
Completeness (_total/calc) 90.21%
PLAT022_ALERT_3_C Ratio Unique / Expected Reflections too Low .... 0.90
PLAT127_ALERT_1_C Implicit Hall Symbol Inconsistent with Expl .... P -1
PLAT220_ALERT_2_C Large Non-Solvent C Ueq(max)/Ueq(min) ... 3.38 Ratio
PLAT230_ALERT_2_C Hirshfeld Test Diff for C4 - C5 = 5.84 su
PLAT355_ALERT_3_C Long O-H Bond (0.82A) O2 - H1 = 1.06 Ang.
Alert level C
REFLT03_ALERT_3_C Reflection count < 95% complete
From the CIF: _diffrn_reflns_theta_max 68.19
From the CIF: _diffrn_reflns_theta_full 0.00
From the CIF: _reflns_number_total 1429
TEST2: Reflns within _diffrn_reflns_theta_max
Count of symmetry unique reflns 1584
Completeness (_total/calc) 90.21%
PLAT022_ALERT_3_C Ratio Unique / Expected Reflections too Low .... 0.90
PLAT127_ALERT_1_C Implicit Hall Symbol Inconsistent with Expl .... P -1
PLAT220_ALERT_2_C Large Non-Solvent C Ueq(max)/Ueq(min) ... 3.38 Ratio
PLAT230_ALERT_2_C Hirshfeld Test Diff for C4 - C5 = 5.84 su
PLAT355_ALERT_3_C Long O-H Bond (0.82A) O2 - H1 = 1.06 Ang.
1 ALERT level A = In general: serious problem
0 ALERT level B = Potentially serious problem
6 ALERT level C = Check and explain
0 ALERT level G = General alerts; check
1 ALERT type 1 CIF construction/syntax error, inconsistent or missing data
2 ALERT type 2 Indicator that the structure model may be wrong or deficient
4 ALERT type 3 Indicator that the structure quality may be low
0 ALERT type 4 Improvement, methodology, query or suggestion
Data collection: PROCESS-AUTO (Rigaku, 1998); cell refinement: PROCESS-AUTO; data reduction: TEXSAN (Molecular Structure Corporation, 2001); program(s) used to solve structure: SHELXS86 (Sheldrick, 1985); program(s) used to refine structure: TEXSAN; molecular graphics: ORTEPIII (Burnett & Johnson, 1996); software used to prepare material for publication: TEXSAN.
Crystal data top
| C18H24N2O4 | Z = 1 |
| Mr = 332.40 | F(000) = 178.0 |
| Triclinic, P1 | Dx = 1.275 Mg m−3 |
| Hall symbol: P -1 | Cu Kα radiation, λ = 1.5418 Å |
| a = 5.2257 (7) Å | Cell parameters from 3066 reflections |
| b = 6.8930 (9) Å | θ = 3.7–68.1° |
| c = 12.162 (1) Å | µ = 0.74 mm−1 |
| α = 97.72 (1)° | T = 93 K |
| β = 91.93 (1)° | Platelet, black |
| γ = 93.66 (1)° | 0.35 × 0.20 × 0.03 mm |
| V = 432.83 (9) Å3 | |
Data collection top
Rigaku RAXIS-RAPID Imaging Plate
diffractometer | 775 reflections with F2 > 2σ(F2) |
| Detector resolution: 10.00 pixels mm-1 | Rint = 0.038 |
| 48 frames, delta ω = 15 deg scans | θmax = 68.2° |
Absorption correction: multi-scan
(Higashi, 1995) | h = −6→6 |
| Tmin = 0.738, Tmax = 0.978 | k = −8→8 |
| 3858 measured reflections | l = −14→14 |
| 1429 independent reflections | |
Refinement top
| Refinement on F2 | H-atom parameters not refined |
| R[F2 > 2σ(F2)] = 0.047 | w = 1/[σ2(Fo2) + (0.05[Max(Fo2,0) + 2Fc2]/3)2] |
| wR(F2) = 0.117 | (Δ/σ)max = 0.016 |
| S = 1.06 | Δρmax = 0.26 e Å−3 |
| 1421 reflections | Δρmin = −0.31 e Å−3 |
| 112 parameters | |
Special details top
Refinement. Refinement using reflections with F2 > -3.0 σ(F2). The
weighted R-factor (wR) and goodness of fit (S) are based
on F2. R-factor (gt) are based on F. The threshold
expression of F2 > 2.0 σ(F2) is used only for calculating
R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| O1 | 0.1064 (3) | 0.7748 (2) | 0.9472 (1) | 0.0563 (5) | |
| O2 | 0.9064 (3) | 0.5406 (2) | 1.1846 (1) | 0.0604 (5) | |
| N1 | 0.3045 (3) | 1.0093 (3) | 1.1258 (1) | 0.0515 (5) | |
| C1 | 0.7098 (4) | 0.5954 (3) | 1.1259 (2) | 0.0466 (6) | |
| C2 | 0.6123 (4) | 0.7753 (3) | 1.1603 (2) | 0.0506 (7) | |
| C3 | 0.4082 (4) | 0.8379 (3) | 1.1003 (2) | 0.0463 (6) | |
| C4 | 0.2937 (4) | 0.7116 (3) | 1.0013 (2) | 0.0455 (6) | |
| C5 | 0.3957 (3) | 0.5312 (3) | 0.9694 (2) | 0.0418 (6) | |
| C6 | 0.3796 (4) | 1.1551 (3) | 1.2203 (2) | 0.0536 (7) | |
| C7 | 0.2453 (5) | 1.1212 (3) | 1.3246 (2) | 0.0681 (8) | |
| C8 | 0.3248 (6) | 1.2774 (4) | 1.4211 (2) | 0.097 (1) | |
| C9 | 0.2008 (7) | 1.2514 (5) | 1.5269 (2) | 0.141 (1) | |
| H1 | 0.9361 | 0.4043 | 1.1369 | 0.1101* | |
| H2 | 0.6846 | 0.8559 | 1.2248 | 0.0607* | |
| H3 | 0.1692 | 1.0381 | 1.0777 | 0.0618* | |
| H4 | 0.5593 | 1.1534 | 1.2344 | 0.0643* | |
| H5 | 0.3411 | 1.2802 | 1.2022 | 0.0643* | |
| H6 | 0.2858 | 0.9973 | 1.3438 | 0.0818* | |
| H7 | 0.0654 | 1.1212 | 1.3107 | 0.0818* | |
| H8 | 0.5053 | 1.2776 | 1.4336 | 0.1166* | |
| H9 | 0.2831 | 1.4006 | 1.4012 | 0.1166* | |
| H10 | 0.2470 | 1.1317 | 1.5502 | 0.1688* | |
| H11 | 0.0197 | 1.2479 | 1.5158 | 0.1688* | |
| H12 | 0.2565 | 1.3577 | 1.5822 | 0.1688* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| O1 | 0.0558 (10) | 0.055 (1) | 0.0578 (9) | 0.0151 (8) | −0.0072 (8) | 0.0029 (8) |
| O2 | 0.061 (1) | 0.059 (1) | 0.0589 (10) | 0.0158 (8) | −0.0146 (8) | −0.0021 (8) |
| N1 | 0.059 (1) | 0.045 (1) | 0.049 (1) | 0.0101 (9) | −0.0041 (9) | −0.0021 (8) |
| C1 | 0.048 (1) | 0.044 (1) | 0.047 (1) | 0.005 (1) | 0.001 (1) | 0.005 (1) |
| C2 | 0.057 (1) | 0.047 (1) | 0.046 (1) | 0.003 (1) | −0.001 (1) | −0.001 (1) |
| C3 | 0.051 (1) | 0.041 (1) | 0.047 (1) | 0.004 (1) | 0.008 (1) | 0.003 (1) |
| C4 | 0.045 (1) | 0.046 (1) | 0.045 (1) | 0.002 (1) | 0.0043 (10) | 0.004 (1) |
| C5 | 0.042 (1) | 0.039 (1) | 0.044 (1) | 0.005 (1) | 0.0027 (9) | 0.0044 (9) |
| C6 | 0.058 (1) | 0.046 (1) | 0.054 (1) | 0.006 (1) | 0.001 (1) | −0.001 (1) |
| C7 | 0.077 (2) | 0.064 (2) | 0.059 (1) | −0.004 (1) | 0.007 (1) | −0.003 (1) |
| C8 | 0.126 (2) | 0.095 (2) | 0.059 (2) | −0.024 (2) | 0.013 (2) | −0.018 (2) |
| C9 | 0.196 (4) | 0.144 (3) | 0.067 (2) | −0.050 (3) | 0.030 (2) | −0.019 (2) |
Geometric parameters (Å, º) top
| O1—C4 | 1.292 (2) | C6—C7 | 1.509 (3) |
| O2—C1 | 1.334 (2) | C6—H4 | 0.950 |
| O2—H1 | 1.059 | C6—H5 | 0.950 |
| N1—C3 | 1.334 (2) | C7—C8 | 1.509 (3) |
| N1—C6 | 1.447 (3) | C7—H6 | 0.950 |
| N1—H3 | 0.950 | C7—H7 | 0.950 |
| C1—C2 | 1.388 (3) | C8—C9 | 1.487 (4) |
| C1—C5i | 1.426 (3) | C8—H8 | 0.950 |
| C2—C3 | 1.395 (3) | C8—H9 | 0.950 |
| C2—H2 | 0.950 | C9—H10 | 0.951 |
| C3—C4 | 1.474 (3) | C9—H11 | 0.950 |
| C4—C5 | 1.396 (3) | C9—H12 | 0.950 |
| C5—C5i | 1.421 (4) | | |
| | | |
| O1···N1ii | 2.872 (2) | N1···H1vii | 3.424 |
| O1···C6ii | 3.302 (3) | N1···C3iv | 3.433 (3) |
| O1···C1iii | 3.354 (2) | C1···C4vi | 3.546 (3) |
| O1···O1ii | 3.463 (3) | C4···H1iii | 3.367 |
| O1···C6iv | 3.473 (3) | C4···C6iv | 3.434 (3) |
| O1···C5v | 3.523 (2) | C5···H1iii | 3.350 |
| O1···N1iv | 3.526 (2) | C5···C6iv | 3.546 (3) |
| O2···C4vi | 3.341 (2) | C6···H1vii | 3.189 |
| O2···C3vi | 3.491 (2) | C6···H1viii | 3.531 |
| N1···C4iv | 3.317 (2) | | |
| | | |
| C1—O2—H1 | 99.8 | C7—C6—H4 | 108.4 |
| C3—N1—C6 | 125.8 (2) | C7—C6—H5 | 108.4 |
| C3—N1—H3 | 117.1 | H4—C6—H5 | 109.5 |
| C6—N1—H3 | 117.1 | C6—C7—C8 | 112.3 (2) |
| O2—C1—C2 | 118.6 (2) | C6—C7—H6 | 108.8 |
| O2—C1—C5i | 119.7 (2) | C6—C7—H7 | 108.8 |
| C2—C1—C5i | 121.7 (2) | C8—C7—H6 | 108.7 |
| C1—C2—C3 | 120.1 (2) | C8—C7—H7 | 108.8 |
| C1—C2—H2 | 120.0 | H6—C7—H7 | 109.5 |
| C3—C2—H2 | 120.0 | C7—C8—C9 | 114.9 (2) |
| N1—C3—C2 | 124.4 (2) | C7—C8—H8 | 108.1 |
| N1—C3—C4 | 115.7 (2) | C7—C8—H9 | 108.1 |
| C2—C3—C4 | 119.9 (2) | C9—C8—H8 | 108.1 |
| O1—C4—C3 | 118.2 (2) | C9—C8—H9 | 108.1 |
| O1—C4—C5 | 123.3 (2) | H8—C8—H9 | 109.5 |
| C3—C4—C5 | 118.5 (2) | C8—C9—H10 | 109.5 |
| C1i—C5—C4 | 120.2 (2) | C8—C9—H11 | 109.5 |
| C1i—C5—C5i | 118.5 (2) | C8—C9—H12 | 109.5 |
| C4—C5—C5i | 121.3 (2) | H10—C9—H11 | 109.4 |
| N1—C6—C7 | 113.6 (2) | H10—C9—H12 | 109.4 |
| N1—C6—H4 | 108.4 | H11—C9—H12 | 109.5 |
| N1—C6—H5 | 108.4 | | |
| Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) −x, −y+2, −z+2; (iii) x−1, y, z; (iv) −x+1, −y+2, −z+2; (v) −x, −y+1, −z+2; (vi) x+1, y, z; (vii) x−1, y+1, z; (viii) x, y+1, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H1···O1i | 1.06 | 1.49 | 2.517 (2) | 161 |
| N1—H3···O1 | 0.95 | 2.24 | 2.664 (2) | 106 |
| N1—H3···O1ii | 0.95 | 2.04 | 2.871 (2) | 146 |
| Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) −x, −y+2, −z+2. |
 O hydrogen bonds to form a ribbon structure along [1
O hydrogen bonds to form a ribbon structure along [1![[sigma]](//journals.iucr.org/logos/entities/sigma_rmgif.gif) (C-C) = 0.003 Å
(C-C) = 0.003 ÅAlert level A PLAT029_ALERT_3_A _diffrn_measured_fraction_theta_full Low ....... 0.91
Alert level C REFLT03_ALERT_3_C Reflection count < 95% complete From the CIF: _diffrn_reflns_theta_max 68.19 From the CIF: _diffrn_reflns_theta_full 0.00 From the CIF: _reflns_number_total 1429 TEST2: Reflns within _diffrn_reflns_theta_max Count of symmetry unique reflns 1584 Completeness (_total/calc) 90.21% PLAT022_ALERT_3_C Ratio Unique / Expected Reflections too Low .... 0.90 PLAT127_ALERT_1_C Implicit Hall Symbol Inconsistent with Expl .... P -1 PLAT220_ALERT_2_C Large Non-Solvent C Ueq(max)/Ueq(min) ... 3.38 Ratio PLAT230_ALERT_2_C Hirshfeld Test Diff for C4 - C5 = 5.84 su PLAT355_ALERT_3_C Long O-H Bond (0.82A) O2 - H1 = 1.06 Ang.

 journal menu
journal menu










 Alert level A
Alert level A
 Alert level C
Alert level C



