issue contents
October 2001 issue
Molecular replacement and its relatives
Proceedings of the CCP4 study weekend
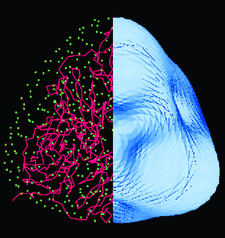
Cover illustration: Molecular shape of AxNiR obtained directly from solution scattering and located in the unit cell by using the FSEARCH program (p. 1410). Dr J. G. Grossmann is thanked for the help with the figures.
research papers
Open  access
access
 access
accessA discussion of the elementary properties of rotation matrices and their represention as polar or Eulerian angles.
Open  access
access
 access
accessA review is given of the mathematical procedures required for a molecular-replacement structure determination.
Open  access
access
 access
accessAn account is given of the molecular replacement method as implemented in the package AMoRe.
Open  access
access
 access
accessThe use of likelihood, particularly a multivariate likelihood function for multiple molecular-replacement models, allows difficult molecular-replacement problems to be solved more easily.
Open  access
access
 access
accessThe locked rotation and translation functions are a powerful way of taking advantage of the presence of non-crystallographic symmetry in a structure determination by the molecular-replacement method.
Open  access
access
 access
accessThe principles and the implementation of the molecular-replacement procedures in CNS are reviewed.
Open  access
access
 access
accessTwo cases of successful molecular replacement using NMR trial models are presented. One is the crystal structure of the E. coli colicin immunity protein Im7; the other is the previously unreported crystal structure of the carboxy-terminal SH2 domain from the p85α subunit of human phosphatidylinositol 3-OH kinase complexed to a PDGF receptor-derived specificity peptide.
Open  access
access
 access
accessThis review summarizes the steps required to exploit electron-microscopy images as models for molecular replacement, indicating some of the associated practical problems.
Open  access
access
 access
accessThe program FSEARCH can be used for a molecular-replacement solution, given a predetermined envelope from any source, such as solution scattering.
Open  access
access
 access
accessThe paper describes the structure solution of UDP-galactopyranomutase. The structure solution relied on the placement of experimental density into a new crystal form.
Open  access
access
 access
accessMathematical data mining is used to generate a representative set of protein fragments to use as search models in molecular replacement.
Open  access
access
 access
accessThe proposal is investigated that protein tertiary structure prediction methods and threading methods in particular might be applied to the problem of solving a protein structure by X-ray crystallography.
Open  access
access
 access
accessFast Fourier feature recognition is one approach to the automatic identification of protein-like features from crystallographic data. This approach has been implemented in a computationally efficient manner and refined to work with inferior data.
Open  access
access
 access
accessThe capabilities of the ARP/wARP software for refining and automatically building protein models, starting from molecular-replacement solutions, are presented. Useful protocols are suggested based on worked examples.
Open  access
access
 access
accessThe molecular-replacement method has been extended to locate macromolecular fragments in an electron-density map using a spherically averaged phased translation and a phased rotation function.
Open  access
access
 access
accessAn overview of the use of NMR models to solve crystal structures by molecular replacement is presented with particular emphasis on the preparation of search models. A recently developed protocol is tested and found to offer good results.
Open  access
access
 access
accessA molecular-replacement method is described which simultaneously determines the rotational and translational parameters of all copies of a search model in the asymmetric unit of a target crystal structure.
Open  access
access
 access
accessRapid six-dimensional molecular-replacement searches can be carried out using an evolutionary optimization algorithm. The performance of this algorithm and its dependence on search-model quality and target function are discussed.
Open  access
access
 access
accessThe anomalous signal of S atoms can contribute to many aspects of solving protein structures and should be used routinely. For example, the anomalous scattering of sulfur was essential to solve the structure of trypsin which was initially phased from a single intrinsic Ca2+ ion in an SAS diffraction experiment.


 journal menu
journal menu