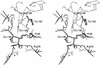issue contents
January 2003 issue
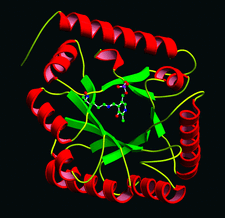
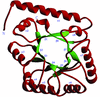
Cover illustration: A representation of the fold of yeast hypothetical protein with PLP and protein ligand Lys49 which is bound to PLP (p. 127).
editorial
Free 

topical reviews
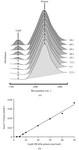
Physicochemical insights into protein crystallization investigated with the Advanced Protein Crystallization Facility are critically reviewed.
research papers
The structure of the N-terminal domain of human FKBP52 was determined using molecular replacement to 2.4 Å. The overall structure is very similar to FKBP12, and the structure–functional relationship of the protein is discussed.
PDB reference: FKBP52, 1n1a
The structure of an enzyme involved in the non-mevalonate pathway of isoprenoid synthesis in the thermophilic bacterium Thermus thermophilus has been solved to high resolution in the presence of substrate and with a substrate analogue.
The calcium bond-valence sum (CBVS) is proposed as a convenient way of identifying metal ions in protein structures.
Open  access
access
 access
accessA method for automated macromolecular main-chain model building is described.
Open  access
access
 access
accessA method for automated macromolecular side-chain model building and for aligning the sequence to the map is described.
The 1.4 Å resolution three-dimensional structure of a homodimeric cholesterol esterase (Candida cylindracea) in complex with an unknown ligand revealed the presence of a C23-chain fatty acid in an elongated enzyme active site.
PDB reference: cholesterol esterase–ligand complex, 1llf, r1llfsf
The phasing of proteins using the anomalous signal from inherent sulfur or halides in the solvent requires the coordinates of an appreciable number of partially occupied or weak sites to be determined. The use of dual-space recycling methods, as implemented in the program SHELXD, to solve such anomalous scatterer substructures in MAD, SAD, SIR and SIRAS experiments has been tested on a previously unknown protein structure.
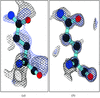
An A-DNA decamer at 1.25 Å resolution shows a novel binding mode of cobalt hexammine ion to DNA duplexes. Analysis of thermal parameters (B factors) indicates that nucleotides involved in abutting crystal packing are thermally more stable than other nucleotdes in the duplex.
PDB reference: d(CCCGATCGGG), 1m77, r1m77sf
NDB reference: d(CCCGATCGGG), ad0026
The 2.0 Å resolution crystal structure of E. coli uridine phosphorylase suggests greater structural similarity to E. coli purine phosphorylase, lending further support to the hypothesis that members of the related but functionally distinct NP-I enzyme class evolved from a common ancestor.
PDB reference: uridine phosphorylase, 1lx7, r1lx7sf
A simple rapid method using infrared spectroscopy for measuring lipid:protein ratios and detergent concentrations in solubilized integral membrane-protein samples has been developed. This analytical tool will aid in the purification and solubilization of membrane proteins that are ideal for crystallization trials.
The crystal structure of E. chrysanthemi L-asparaginase has been refined at 1.0 Å resolution, elucidating the atomic level details of one of the largest asymmetric units studied to date at this level of accuracy.
PDB reference: L-asparaginase, 1o7j, r1o7jsf
Using synchrotron radiation and a CCD detector, X-ray data have been collected at 100 K for the His64Ala mutant of human carbonic anhydrase II complexed with 4-methylimidazole (4-MI) to a maximal 1.05 Å resolution, allowing full anisotropic least-squares refinement.
PDB reference: carbonic anhydrase II, 1moo, r1moosf
The structure of T. aurantiacus xylanase solved to small-molecule accuracy at atomic resolution (1.11 Å) at 293 K and at ultrahigh resolution (0.89 Å) at 100 K provides insights into the plasticity of salt bridges, the temperature-dependent deformation and the water structure of the enzyme, which belongs to the ubiquitous TIM-barrel fold.
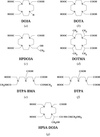
Seven gadolinium complexes are shown to be excellent compounds for the production of heavy-atom derivatives for macromolecular crystallography projects. SAD phasing has been successfully carried out on a series of gadolinium-derivative crystals of two proteins, hen egg-white lysozyme and urate oxidase from A. flavus, leading to high-quality electron-density maps.
structural genomics papers
The crystal structure of YBL036C has been determined by MIRAS and MAD methods.
crystallization papers
Preliminary neutron-diffraction measurements of E. coli trp repressor crystals demonstrate that it will be possible to obtain a full-atom neutron structural model of a DNA-binding protein plus its associated solvent.
Crystallization of family 9 A. acidocaldarius CelA is reported. The crystals are unusual in that they do not bind the dye IZIT in the mother liquor and do not dissolve in the protein-storage buffer or water for several weeks. Collection of a data set to 3.0 Å resolution and initial molecular-replacement searches are reported.
An all-RNA 64-mer hairpin ribozyme has been crystallized in a form suitable for X-ray diffraction analysis at 3.35 Å resolution. The construct contains the U39C substitution, which enhances catalytic efficiency and represents the minimal catalytic motif formed by intermolecular docking of two helix–loop–helix domains. The crystallization strategy and results are presented.
A partial extracellular fragment of a sperm membrane protein YWK-II protein/APPH related to the Alzheimer βA4-amyloid precursor protein has been recloned, expressed in E. coli, crystallized and a complete native data set has been collected at 2.8 Å resolution.
The first crystallographic study, at 2.2 Å resolution, of an enzyme that is involved with both hydrolysis and isomerization of sucrose.
Overexpression, purification and crystallization of T. maritima TruB allowed collection of a complete data set to 2.0 Å resolution.
The GTP-responsive transcriptional repressor CodY from B. subtilis has been crystallized in a number of forms in the presence and absence of ligands. A complete data set to 3.5 Å spacing has been collected.
Good-quality crystals have been obtained of two microbial α-amino-acid ester hydrolases, enzymes of potential use in antibiotic production. One enzyme crystallizes in two space groups, one of which has extremely large unit-cell parameters.
A Jel44 monoclonal Fab fragment specific for E. coli HPr has been purified. Crystals of the Jel44 Fab fragment diffracted to a resolution of 4.6 Å, while crystals of the novel complex of the Jel44 Fab fragment with HPr diffracted to a resolution of 2.5 Å.
Preliminary crystallographic studies of synthetically prepared thymus and activation-regulated chemokine (TARC) in the triclinic, hexagonal and tetragonal systems are described.
Malonyl-CoA decarboxylase of a Rhizobium species was crystallized. This enzyme shares 30% sequence identity with human homology.
The in vivo role and substrate specificity of sulfurtransferase enzymes has been greatly debated. SseA, a 3-mercaptopyruvate:cyanide sulfurtransferase from E. coli, has been crystallized in a tetragonal crystal form suitable for X-ray crystallographic investigations.
The Holliday junction resolving enzyme Hje from S. solfataricus has been crystallized. Preliminary crystallographic analysis indicates a possible difference in dimeric arrangement to the homologous enzyme Hjc.
Crystals of a zinc-containing alcohol dehydrogenase from the hyperthermophilic archaeon A. pernix have been grown by the sitting-drop vapour-diffusion technique from recombinant protein expressed in E. coli and show diffraction to 1.5 Å resolution. The anomalous diffraction from zinc at two wavelengths and twofold NCS averaging have resulted in an interpretable electron-density map.
Crystals of complexes between the radixin FERM domain and the C-terminal regions of NHERF and NHERF2 have been obtained.
CMP-KDO synthetase from H. influenzae was crystallized. X-ray diffraction data were collected to 2.2 Å from an SeMet crystal.
tRNA(m1G37)methyltransferase (TrmD) from H. influenzae was crystallized. X-ray data have been collected to 1.85 Å.
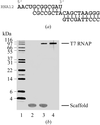
A phage T7 RNA polymerase elongation complex containing 8 bp of RNA–DNA hybrid and 10 bp of downstream DNA with a total molecular mass of ∼110 kDa was crystallized. A complete diffraction data set was collected at 2.6 Å resolution. The structure determination and refinement are in progress.
3-Chlorocatechol 1,2-dioxygenase from the Gram-positive bacterium R. opacus (erythropolis) 1CP, an enzyme involved in the specific cleavage of 3-chlorocatechol, which is an intermediate in the aerobic biodegradation of 2-chlorophenol, has been crystallized.
The kinase-recruitment domain of the PP2C-type phosphatase RsbU from B. subtilis has been crystallized in a form that diffracts to 1.6 Å resolution using synchrotron radiation.
short communications
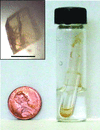
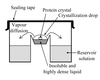
A new method of protein crystallization, the floating-drop vapour-diffusion technique, has been developed.
international union of crystallography
Free 



 journal menu
journal menu