issue contents
August 2008 issue
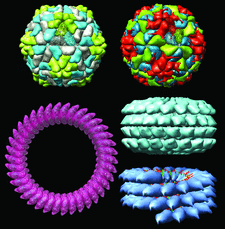
Cover illustration: Examples of recently remediated PDB entries with regular noncrystallographic symmetry (p. 874). Top left: yellow mottle virus (1f2n); top right: rhinovirus (4rhv); bottom left: pneumolysin (2bk1); center right: tobacco mosaic virus coat protein aggregate (1ei7); bottom right: cucumber green mottle mosaic virus (1cgm).
research papers
Crystal structures of urate oxidase from A. globiformis and of its complexes with uric acid, allantoate and 8-azaxanthin demonstrate details of substrate recognition and catalysis.
Improved classification of images from crystallization experiments is obtained using multiple classifiers to combine different feature-extraction methods.
A pattern-recognition-based method for the identification of planar objects in crystallographic electron-density maps is presented. The accuracy of the located centres of the planes is of the order of 0.5 Å.
The structure of the 1918 H1N1 neuraminidase was determined to 1.65 Å from crystals with a lattice-translocation defect using uncorrected, as well as corrected, diffraction data.
PDB reference: 1918 H1N1 neuraminidase, 3cye, r3cyesf
Open  access
access
 access
accessThe short hydrogen bonds in rhamnogalacturonan acetylesterase have been investigated by structure determination of an active-site mutant, 1H NMR spectra and computational methods. Comparisons are made to database statistics. A very short carboxylic acid carboxylate hydrogen bond, buried in the protein, could explain the low-field (18 p.p.m.) 1H NMR signal.
PDB reference: D192N rhamnogalacturonan acetylesterase, 3c1u, r3c1usf
A novel phasing method that uses the charge-flipping algorithm has been used to solve the ab initio structure of biological macromolecules at atomic resolution and to determine heavy-atom or anomalous scattering substructures.
Open  access
access
 access
accessA new data model for PDB entries of viruses and other biological assemblies with regular noncrystallographic symmetry is described.
Structures of grape dihydroflavonol 4-reductase in complex with NADP+ and various flavonols suggest possible inhibition of the enzyme by flavonols. Initial kinetics experiments confirmed the inhibition and showed its competitive character.
Structural insights were obtained into the copper-chelation mechanism in superoxide dismutases.
PDB reference: superoxide dismutase, 2q2l, r2q2lsf
short communications
Open  access
access
 access
accessTwo crystal structures of rhodopsin that were originally described using trigonal symmetry can be interpreted in a hexagonal unit cell with a smaller asymmetric unit.
The crystal-packing and cohesive energies in the structures of two polymorphs of the title tetrapeptide have been analyzed using molecule–molecule energies calculated using the PIXEL method.


 journal menu
journal menu