issue contents
March 2010 issue
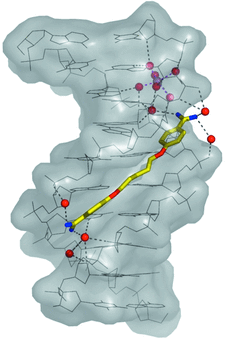
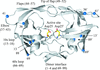
Cover illustration: View of the pentamidine-d(ATATATATAT)2 complex (p. 251). The drug is in the minor groove at the centre of the sequence. The water molecules and one Mg2+ ion found in the minor groove are also shown. Hydrogen bonds are represented as dashed lines. The two ends of the duplex are different. At one end (top) the duplex forms standard Watson-Crick base pairs. At the other end (bottom) a Hoogsteen A·T base pair is present. Note that the Mg2+ ion is at the back of the duplex in this presentation.
research papers
In this study, the crystal structures of the human Hsp70 nucleotide-binding domain (NBD) fragment were determined in the nucleotide-free state and in complex with adenosine 5′-(β,γ-imido)triphosphate (AMPPNP).
The crystal structure of the unbound form of HIV-1 subtype A protease has been determined to 1.7 Å resolution. A detailed structural analysis and comparison of the unbound subtype A, B and C protease structures is presented. The results showed that although no inhibitor is present in the active site, the subtype A protease has flaps in the closed position.
PDB reference: HIV-1 subtype A protease, 3ixo
E. coli alanyl-tRNA synthetase is recalcitrant to crystallization. A group of leucine substitutions has transformed the protein.
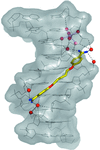
The oligonucleotide d(ATATATATAT) has been crystallized as a coiled coil in the presence of pentamidine. The drug sits in the minor groove of the duplex, but shows a new form of interaction with DNA as a cross-linking agent. The duplex contains both Watson–Crick and Hoogsteen base pairs.
PDB reference: pentamidine–DNA complex, 3ey0
NDB reference: DD0102
The crystal structure of midge larval haemoglobin was determined at five different pH conditions. Comparison of these structures indicated that pH changes result in a swinging movement of the distal Arg residue and a partial shift of the F-helix C-terminus followed by haem tilting in this insect haemoglobin.
PDB references: haemoglobin component V, pH 4.6, 2zwj; pH 5.6, 3a5a; pH 6.5, 3a5b; pH 7.0, 3a5g; pH 9.0, 3a9m
Open  access
access
 access
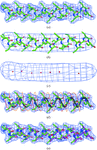
accessA method for rapid model building of α-helices at moderate resolution is presented.
Open  access
access
 access
accessA method for rapid model building of β-sheets at moderate resolution is presented.
Open  access
access
 access
accessA method for rapid chain tracing of polypeptide backbones at moderate resolution is presented.
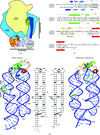
The structures of the human and a crenarchaeal SRP19–SRP RNA complex reveal general features of SRP S-domain organization and allow an update of the SRP-assembly paradigm.
short communications
The solution of a structural protein from the Acidianus two-tailed virus using long-wavelength SAD phasing suggests a systematic approach for the solution of protein crystal structures by taking advantage of the anomalous signal of serendipitously associated chloride ions from the solvent.
PDB reference: Structural protein from the Acidianus two-tailed virus, 3faj
Procedures for heavy-atom site identification by molecular replacement at low resolution and for density modification by multi-crystal averaging using quasi-isomorphous diffraction data are presented. The procedures are based on the case history from the structure determination of H+-ATPase and Na+, K+-ATPase and have been successfully applied to the transporter Mhp1 to reveal general applicability.
Variable packing interaction related to the conformational flexibility within the huntingtin-interacting protein 1 coiled coil domain.
PDB reference: Huntingtin-interacting protein 1 coiled-coil domain, 3i00
Deleting two C-terminal α-helices of the MsbA molecule from E. coli improved its crystallizability and generated new crystal forms of the complex with adenosine 5′-(β,γ-imido)triphosphate that had an outward-facing conformation.


 journal menu
journal menu