issue contents
February 2011 issue
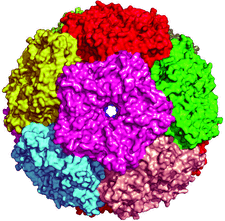
Cover illustration: Surface representation of the icosahedral assembly of lumazine synthase from Salmonella typhimurium, showing each pentamer in a different colour, p. 131.
research papers
The structure of the effector-binding domain of the LysR-type transcription regulator RovM from Y. pseudotuberculosis is reported at 2.4 Å resolution, showing a potential inducer-binding cavity.
PDB reference: effector-binding domain of RovM, 3onm
The 1.86 Å resolution crystal structure of propanediol-utilization (Pdu) microcompartment shell protein PduT from the bacterium Citrobacter freundii is reported.
PDB reference: PduT, 3pac
An oligonucleotide representing a regulatory element of human thymidylate synthase mRNA has been crystallized as a dimer. The structure of the asymmetric dimer has been determined at 1.97 Å resolution.
PDB reference: TS1 dimer, 3mei
NDB reference: TS1 dimer, NA0505
TTHA0988 from T. thermophilus represents a class of protein found almost ubiquitously across bacterial species which is incorrectly annotated as allophanate hydrolase. Three crystal forms of TTHA0988 have been solved which reveal the domain organization and range of motion within this protein family.
Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
In order to study the effects of three amino-acid substitutions found in PAD4SNP, the crystal structure of PAD4SNP has been determined.
Open  access
access
 access
accessX-ray crystal structures of the cyclosporin A analogue E-ISA247 (voclosporin) and its stereoisomer Z-ISA247 bound to cyclophilin A suggest the molecular basis for the differences in their binding affinities and immunosuppressive efficacies.
The structure of XcpWJ has been refined to 1.85 Å resolution and revealed a type IVa pilin fold with an embellished variable antiparallel β-sheet.
PDB reference: XcpW, 3nje
The structure of icosahedral lumazine synthase from S. typhimurium is reported at 3.57 Å resolution.
PDB reference: lumazine synthase, 3mk3
The crystal structure of the trypsin–bovine pancreatic trypsin inhibitor (BPTI) complex was refined using X-ray and neutron data. Both protonation states of the catalytic triad and H/D exchange of the amide H atom of P1′ Ala16 of the scissile peptide bond were observed in the trypsin–BPTI complex.
PDB reference: trypsin–BPTI complex, 3otj


 journal menu
journal menu