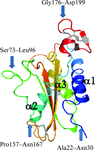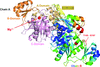issue contents
October 2011 issue
Cover illustration: NADP+ binding in the highly specific isocitrate dehydrogenase from C. glutamicum (p. 856). Specificity is mediated by interactions between potentially positively charged side chains of the enzyme with the negatively charged 2'-phosphate group of the coenzyme, which is ordered mainly at its adenosine nucleoside end.
research papers
MetY is the first reported structure of an O-acetylhomoserine sulfhydrylase that utilizes a protein thiocarboxylate intermediate as the sulfur source in a novel methionine-biosynthetic pathway instead of catalyzing a direct sulfhydrylation reaction.
PDB reference: MetY, 3ri6
Complementary measurements using SONICC and TPE-UVF allow the sensitive and selective detection of protein crystals.
Structural analysis of a truncated soluble domain of human glioma pathogenesis-related protein 1, a membrane protein implicated in the proliferation of aggressive brain cancer, is presented.
The crystal structure of the highly NADP+- and isocitrate-specific monomeric isocitrate dehydrogenase from C. glutamicum in complex with NADP+ and the cofactor Mg2+ shows the holoenzyme in a conformation that is accessible to the substrate. A disrupted substrate-binding site is in close proximity to one of two pairs of hinge residues, suggesting a model of how substrate binding close to the hinge may lead to large-scale conformational changes that appear to be functionally important.
PDB reference: CgIDH-Holo, 3mbc
The crystal structure of N5-carboxyaminoimidazole ribonucleotide synthase from Bacillus anthracis with only an Mg cation provides some insight into the catalytic mechanism of this enzyme and the role of a crucial loop during catalysis.
PDB reference: N5-carboxyaminoimidazole ribonucleotide synthase, 3q2o
These data for the binary complex of PT684 with hDHFR reveal that the 3-carboxypropyl side chain occupies two alternate positions, neither of which interacts with the conserved Arg in the active site. These data also do not confirm the computational model for PT684 binding to hDHFR which was based on the structure of PT653 in Pneumocystis carinii DHFR.
PDB reference: hDHFR complex, 3s7a
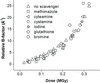
Free-radical scavengers that are known to be effective protectors of proteins in solution are found to increase global radiation damage to protein crystals. Protective mechanisms may become deleterious in the protein-dense environment of a crystal.
X-ray structure of wild-type Chlamydomonas reinhardtii αCA1, matured by post-translational modifications of peptide cleavage and N-glycosylations, has been determined at 1.88 Å resolution.
PDB reference: Cr-αCA1, 3b1b
short communications
A simple method for the mounting of crystals in the absence of mother liquor is presented. The technique has been tested on a number of systems and may be generally applicable, thereby obviating the need for extensive testing of cryoprotectant solutions.


 journal menu
journal menu