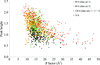issue contents
January 2012 issue
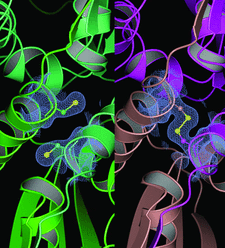
Cover illustration: Bach2 is a POZ-domain transcriptional repressor that regulates B-cell differentiation and oxidative stress responses. Bach2 has been implicated in several human malignancies, and the X-ray crystal structure reported here (p. 26) will be important for the design of therapeutics. The structure revealed an inter-subunit disulfide bond that may be relevant in the regulation of oxidative stress responses.
research papers
A fully automatic and efficient procedure to find α-helices in the Patterson map is described and extensively applied to test structures.
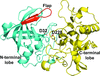
Crystal structure determination, statistical analysis of all the available crystal structures and molecular dynamics simulations studies of BACE1 revealed that the multiple conformations assumed by the flap in its active site are influenced by the presence, position and orientation of a bound inhibitor as well as by the crystal packing.
The POZ-domain transcriptional repressor Bach2 is regulated by oxidative stress and is involved in some human malignancies. The reported crystal structure will be important for the design of cancer therapeutics.
The 1.75 Å resolution crystallographic structure of perdeuterated rubredoxin refined against neutron data collected in just 14 h revealed 269 D atoms without ambiguity.
Open  access
access
 access
accessFine φ-slicing substantially improves scaling statistics and anomalous signal for diffraction data collection with hybrid pixel detectors.
A systematic approach by the molecular-replacement method with a dimeric search model using the program Phaser led to an unexpected pentameric structure of the NSP4:95–146 region of the ST3 strain of rotavirus, in contrast to the previously reported tetramers.
PDB reference: rotavirus NSP4, 3miw
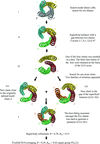
New phasing opportunities arise when crystals are grown from a racemic or quasi-racemic mixture of a protein molecule and its synthetic mirror image enantiomer. In this study, good phases are obtained from single wavelength (SAD) data by exploiting the nearly centrosymmetric nature of a quasi-racemic crystal.
The stabilizing osmolyte trimethylamine N-oxide (TMAO) is shown to be an efficient primary precipitant for protein crystal growth. In addition to TMAO, two other methylamine osmolytes, sarcosine and betaine, are shown to be effective cryoprotective agents for protein crystal cooling.
PDB references: trypsin, TMAO-grown, tetragonal, 3t28; TMAO-grown, trigonal, 3t29; TMAO-grown, orthorhombic, 3t25; in the presence of sarcosine, 3t26; in the presence of betaine, 3t27; insulin, TMAO-grown, 3t2a; thermolysin, in the presence of TMAO, 3t2h; in the presence of sarcosine, 3t2i; in the presence of betaine, 3t2j
Open  access
access
 access
accessThe tertiary structure and kinetic properties of alanine racemase from Staphylococcus aureus are described and compared to other related alanine racemase structures.
PDB reference: alanine racemase, 4a3q
book reviews
Free 



 journal menu
journal menu