issue contents
October 2015 issue
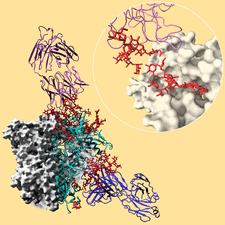
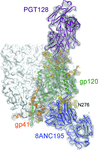
Cover illustration: Broadly neutralizing antibodies PGT128 (purple ribbon) and 8ANC195 (blue ribbon) interact with the glycan shield (red ball-and-stick) of the HIV-1 envelope glycoprotein trimer (Kong et al., p. 2099). Antibody interactions with one protomer of the HIV-1 trimer are shown. Each protomer (turquoise ribbon) is comprised of gp120 and gp41. The other two protomers of the trimer are displayed as white molecular surfaces. The inset zooms in on the interaction between PGT128 (purple ribbon) and gp120 (white molecular surface). PGT128 allosterically interacts with the glycan shield of the HIV-1 envelope glycoprotein trimer. PGT128 contacts an N-linked glycan at position Asn301 on the HIV-1 trimer, which then alters the conformation of the N-linked glycan at position Asn262, which differs from the unliganded Asn262 glycan conformation. The information derived from this crystal structure is being used for design of HIV-1 vaccine immunogens. This illustration was generated by Christina Corbaci and Leopold Kong.
research papers
 access
access

 access
access



 journal menu
journal menu