issue contents
July 2021 issue
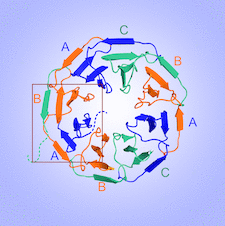
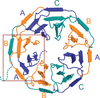
Cover illustration: Crystal structure of Scone: pseudosymmetric folding of a symmetric designer protein [Mylemans et al. (2021), Acta Cryst. D77, 933–942]. Here, attempts were made to engineer a ninefold-symmetric β-propeller protein into a threefold-symmetric nine-bladed propeller using computational design. Two nine-bladed propeller proteins were designed, named Scone-E and Scone-R. Crystallography, however, revealed the structure of both designs to adopt an eightfold conformation with distorted termini, leading to a pseudo-symmetric protein.
topical reviews
Open  access
access
 access
accessThis article lists the basic crystallographic equations underlying molecular-replacement software. The prospects for future developments are considered, and a brief account of past progress is included.
research papers
Open  access
access
 access
accessAn intensity-based likelihood method is provided to estimate scattering from an anomalous substructure considering the effect of measurement errors in Bijvoet pairs and the correlations between these errors.
The structure of PSK, a novel antimicrobial peptide identified in Chrysomya megacephala larvae, was determined by X-ray crystallography and small-angle X-ray scattering, which revealed a distinct fold compared with published homologous peptides.
PDB reference: PSK, 7dtl
Open  access
access
 access
accessParD2 forms an open oligomer in solution, the size of which is limited by the presence of an intrinsically disordered tail. In the absence of this tail, ParD2 forms a circular hexadecamer.
PDB reference: N-terminal domain of ParD2, 7b22
Open  access
access
 access
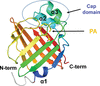
accessThe crystal structure of the ferredoxin reductase component of carbazole 1,9a-dioxygenase from Janthinobacterium sp. J3 was determined at 2.40 Å resolution.
Using computational protein design, a symmetric protein was designed; however, its crystal structures revealed a different pseudosymmetric architecture. Furthermore, one variant was only able to crystallize upon the addition of a polyoxometalate which links individual proteins in the crystal packing.
PDB references: Scone-R, 7awy; Scone-E, 7awz; Scone-E + STA, form A, 7ax0; Scone-E + STA, form B, 7ax2
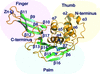
Coronaviruses encode a papain-like protease (PLP) which is responsible for antagonizing the innate immune response; here, the structure of the first known Alphacoronavirus PLP bound to ubiquitin, its preferred substrate, is reported.
PDB reference: PEDV papain-like protease 2 bound to ubiquitin, 7mc9
Based on the crystal structure of human fatty acid-binding protein FABP7 in complex with palmitic acid, the recognition of saturated fatty acids by FABP7 is discussed.
PDB reference: human FABP7 complexed with palmitic acid, 7e25
An automated particle-picking method for cryo-EM and its evaluation using precision TEM simulation based on a multi-slice method are described.
addenda and errata
Open  access
access
 access
access

 journal menu
journal menu