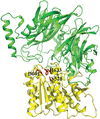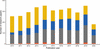issue contents
December 2021 issue

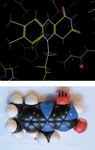
Cover illustration: The validation of automatically generated stereochemical restraints highlights the limitations of using such restraints and the fragility of creating ideal geometry – one of the things Roversi & Tronrud `hate' about refinement [Roversi & Tronrud (2021), Acta Cryst. D77, 1497–1515].
editorial
Open  access
access
 access
accessThe editors discuss the submission of structural biology data.
CCP4
Open  access
access
 access
accessA 3D cellular imaging platform developed at beamline B24 at Diamond Light Source has been used to identify cellular features such as filamentous actin in mammalian cells. This approach enabled virtual same-sample imaging of structures captured on separate microscopes through rigid transformation of 3D data in silico, bypassing the need for additional sample processing and ensuring artefact-free data correlation.
Open  access
access
 access
accessA standalone system, called PDB-Dev, has been developed for archiving integrative structures and making them publicly available. The paper describes the data standards, the software tools and the various components of the PDB-Dev data-collection, processing and archiving infrastructure.
Open  access
access
 access
accessTen goals for the future development of macromolecular refinement programs are described.
research papers
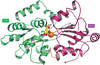
MctA is an Mg2+-complexed citrate-binding protein from a Gram-negative bacterium that belongs to the ABC transporter superfamily. Comparison of the crystal structures of wild-type and mutant MctA proteins suggest a gating mechanism of substrate entry following an `asymmetric domain movement' mechanism of substrate binding.
PDB references: MctA, endogenously bound to Mg2+–citrate complex, form I, 7f6e; form II, 7f6f; endogenously bound to citrate, 7f6k; S26A mutant, endogenously bound to Mg2+–citrate complex, 7f6n; S26A mutant, endogenously bound to Mn2+–citrate complex, 7f6o; D28A mutant, endogenously bound to citrate, 7f6p; S79A mutant, apo state, 7f6q; 164A mutant, apo state, 7f6r; T199A mutant, apo state, 7f6s; Y221F mutant, apo state, 7f6t; Y221A mutant, apo state, 7f6u
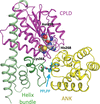
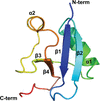
Legionella pneumophila protein effector LegA15/AnkD contains an ankyrin-repeat domain, a cysteine protease-like domain with His268–Asn290–Cys361 putative catalytic triad, and a helix-bundle domain. LegA15/AnkD shows structural similarity to another effector, LegA3/AnkH, but they localize to different cellular compartment within the host.
PDB reference: Legionella pneumophila protein effector LegA15/AnkD, 7kj6
Open  access
access
 access
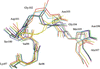
accessThe CIDE domain was initially identified in apoptotic nucleases and now forms a highly conserved family with diverse functions ranging from cell death to lipid metabolism. Based on structural determination of the DREP3 domain, it is suggested that the head-to-tail helical filament structure might be a unified mechanism of CIDE-domain assembly and represents a critical higher-order scaffolding structure that is important for the function of CIDE-domain-containing proteins in DNA fragmentation and lipid-droplet fusion.
PDB reference: DREP3, 7v6e
Extensive biochemical assays revealed that a putative peptidase S9Cfn from Fusobacterium nucleatum is a carboxypeptidase rather than an aminopeptidase as previously noted. Combined with the 2.6 Å tetramer structure of S9Cfn, key residues for substrate binding and catalysis, as well as the underlying mechanism of the catalytic cycle was revealed for S9C peptidase family.
PDB reference: carboxypeptidase from Fusobacterium nucleatum, 7ep9
The X-ray structure of BbgIII, a multidomain β-galactosidase from B. bifidum, determined using an intact protein corresponding to a gene construct of eight domains is reported.
PDB reference: BbgIII from Bifidobacterium bifidum, 7nit
Open  access
access
 access
accessA study of in vitro refolding and isotope effects on protein structure, activity and stability shows that different folding dynamics can lead to important changes in protein properties.
PDB reference: hydrogenated refolded hen egg-white lysozyme, 7p6m
Open  access
access
 access
accessA machine-learning model was used to predict the performance of four crystallographic model-building pipelines (ARP/wARP, Buccaneer, Phenix AutoBuild and SHELXE) and their combinations.
The new CAB automatic model-building program is described and compared with other automated model-building techniques.
The crystal structure of the A. muciniphila arylsulfatase AmAS is reported and some structural regularity of substrate binding and specificity was noted. Insights into the catalytic mechanism of mucin-desulfating sulfatases in A. muciniphila are also provided based on this regularity.


 journal menu
journal menu