issue contents
February 2023 issue
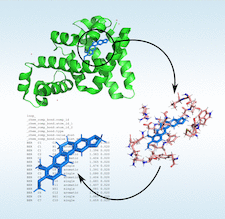
Cover illustration: The Quantum Mechanical Restraints (QMR) procedure generates in situ ligand restraints based on a QM minimization of the ligand in the protein binding pocket [Liebschner et al. (2023), Acta Cryst. D79, 100–110]. The QM procedure is applied to a selection of the protein around the ligand. This illustration uses the example of ligand BER in PDB entry 3vw2.
editorial
Free 

Celebrating 30 years of Acta D and looking forward to the future of structural biology.
scientific commentaries
Free 

The template tables for small-angle scattering data have been updated by Trewhella et al. [(2023), Acta Cryst. D79 122–132].
CCP4
Open  access
access
 access
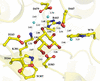
accessIn situ quantum-mechanical optimization of ligand structures provides accurate ligand restraints for macromolecular refinement.
CCP-EM
Open  access
access
 access
accessSARS-CoV-2 spike protein comprises a conserved hydrophobic pocket that binds linoleic acid (LA). LA supplementation interferes with viral entry into the cell and replication, and thus could be used as an antiviral.
research papers
Open  access
access
 access
accessAn updated template reporting table based on the 2017 publication guidelines for biomolecular SAS and 3D modelling is presented that includes standard descriptions for proteins, glycosylated proteins, DNA and RNA, and some reorganization of the data to improve readability and interpretation. A specialized template has also been developed for reporting SAS contrast-variation data and models that incorporates the additional reporting requirements for these more complicated experiments.
Open  access
access
 access
accessThe crystal structure of the self-complementary d(CGCGCG)2 Z-DNA duplex in complex with cadaverinium and potassium cations was solved at ultrahigh resolution. The oligonucleotide used for crystallization contained the enantiomeric 2′-deoxy-L-ribose instead of its natural D-enantiomer, and thus the Z-DNA duplex is right-handed.
Open  access
access
 access
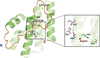
accessAn updated structure of adeno-associated virus serotype 4 (AAV4) is presented with a comparison to receptor-bound structures (AAV2 and AAV5) to predict why AAV4 does not bind to the adeno-associated virus receptor.
EMDB reference: adeno-associated virus serotype 4, EMD-25903
PDB reference: adeno-associated virus serotype 4, 7thr
Open  access
access
 access
accessThe alkaline α-galactosidase AtAkαGal3 from Arabidopsis exhibits a dual function where it can synthesize stachyose using raffinose, instead of galactinol, as the galactose donor. Crystal structures of complexes of AtAkαGal3 and its D383A mutant with various substrates and products, including galactose, galactinol, raffinose, stachyose and sucrose, reveal four complete subsites (–1 to +3) and a new secondary product-binding site, providing the first representative structure of an alkaline α-galactosidase and a model for the raffinose family of oligosaccharide synthases.
Open  access
access
 access
accessThe atomic structure of a polyglycine hydrolase solved using a RoseTTAFold model-assisted molecular-replacement method reveals a two-domain enzyme. The structure reveals a novel tertiary fold and a similarity to β-lactamases.
PDB reference: Fvan-cmp, 7tpu
Open  access
access
 access
accessThe crystallographic structure of the DnaB·DciA complex from Vibrio cholerae reveals that the various helicase loaders share the same binding site on the bacterial replicative helicase, albeit with differences in their loading mechanisms.
PDB reference: VcDnaB·VcDciA, 8a3v
A novel marine bacterial secretory phospholipase A2 (Sm-sPLA2) was characterized as being efficient in liposome biotransformation at high temperatures. The high-resolution crystal structure and mutagenesis studies revealed the determinant of the thermostability of Sm-sPLA2.
PDB reference: secretory phospholipase A2 from Sciscionella marina, 7ygk

 journal menu
journal menu