issue contents
August 2023 issue
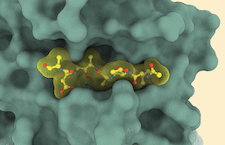
Cover illustration: Crystal structure of PvS1Cat-Tryps (depicted as a green surface) in complex with the inhibitor MAM-117 (yellow) [Martinez et al. (2023), Acta Cryst. D79, 721–734]. The emergence of multi-resistant Plasmodium sp. parasites requires the identification of new antimalarials. Subtilisin-like protease 1 (SUB1) belongs to a new generation of drug targets because it plays a crucial role during egress of the parasite from infected host cells at different stages of its life cycle. The high-resolution structure of Plasmodium vivax SUB1, produced under controlled conditions and complexed with a substrate-derived pseudo-peptide inhibitor, is shown here.
letters to the editor
Open  access
access
 access
accessA descriptor with a standard format and parameters is introduced for the large-area fixed targets used in serial crystallography. It is proposed that any fixed target used and developed by different facilities or groups could have its own version of this descriptor, facilitating their wider use and exchange.
CCP4
Open  access
access
 access
accessRaynals is an online, user-friendly, free (for academia) and advanced tool for the analysis of single-angle dynamic light-scattering data. Estimation of the size distribution is performed through the Tikhonov–Phillips regularization.
Open  access
access
 access
accessA new protein structure validation method using hydrogen-bonding parameters is described.
research papers
Open  access
access
 access
accessThe expression, characterization and structures of CeuE homologues from two thermophilic bacteria, Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius, are described together with their ligand binding. The proteins show enhanced thermostability and resistance to organic chemicals; consequently, these thermophilic homologues offer advantages in the development of artificial metalloenzymes using the CeuE family.
Open  access
access
 access
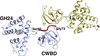
accessModule walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases
The identification, characterization and X-ray structure of a novel fungal GH24 muramidase from Trichophaea saccata is described in which an SH3-like cell-wall-binding domain was identified by structure comparisons in addition to its catalytic domain. A domain-walking approach was then used to identify a group of fungal muramidases that belong to a new GH family containing homologous SH3-like cell-wall-binding modules, and X-ray structures of the independent catalytic and SH3-like domains of three of them are reported.
PDB references: KsGH184, 8b2e; TsCWBD–triglycine complex, 8b2f; PvCWBD, 8b2g; TtGH184, 8b2h; TsCWBD-GH24, 8b2s
The first high-resolution 3D structure of Plasmodium vivax SUB1 complexed with a substrate-derived pseudo-peptide inhibitor is described. Subtle conformational changes in the enzyme–inhibitor complex define new pharmacophores for iterative cycles of structure-driven synthesis to accelerate the discovery of optimized SUB1 inhibitors that could represent a new generation of antimalarial candidates.
Open  access
access
 access
accessThe first structures of Candida auris dihydrofolate reductase at near-atomic resolution are described, including apo and holo forms and complexes with two antifolate drugs: pyrimethamine and cycloguanil. C. auris is a highly significant globally emerging and resistant fungal pathogen and there is a great demand for research and progress towards novel therapeutics.
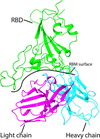
An E77 antibody with high binding affinity for the receptor-binding domain (RBD) of the prototype and Delta variant SARS-CoV-2 spike proteins was identified. The RBD–E77 Fab complex structure shows that the E77 antibody mimics the binding of angiotensin-converting enzyme 2 to RBD and that the N501Y mutation in variants of concern abolishes the binding potency of E77 to RBD.
EMDB reference: RBD–E77 Fab, EMD-35369
PDB reference: RBD–E77 Fab, 8idn
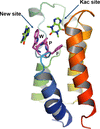
High-resolution crystal structures of members of the purine class of FDA-approved drugs and phytochemical alkaloids complexed with hBRD2 bromodomains BD1 and BD2 revealed a new binding site by stacking the compounds against the WPF shelf of BD1 and BD2 in addition to the known acetyl-lysine binding site.
PDB references: BD1, complex with acyclovir, 7vrh; complex with theophylline, 7vrk; complex with theobromine, 7vro; complex with doxophylline, 7vrz; complex with methylxanthine, 7vsf; BD2, complex with acyclovir, 7vri; complex with theophylline, 7vrm; complex with theobromine, 7vrq; complex with doxophylline, 7vs0; complex with methylxanthine, 7vs1
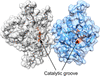
The two L-asparaginases ReAIV and ReAV encoded by the symbiotic bacterium R. etli were previously provisionally assigned to the same structural class 3. Here, it is shown that despite their low sequence identity, the constitutive enzyme ReAIV has the same architecture and active site as the inducible ReAV protein, although its kinetic characteristics are not identical to those of ReAV.

 journal menu
journal menu