issue contents
February 2024 issue
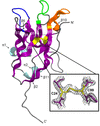
Cover illustration: Time-resolved structural changes in an LOV domain (formation, then disruption of a thioether bond between a cysteine residue and the FMN chromophore) over four orders of time magnitude [Caramello & Royant (2024), Acta Cryst. D80, 60–79]. The sequence starts with the dark state (grey), which then proceeds to a photostationary equilibrium (green) with the light state under continuous illumination (light blue in timeline). After illumination has been stopped, the light state relaxes back to a slightly different dark state (red), concomitant to a change in the crystallographic space group.
CCP4
Open  access
access
 access
accessFrom femtoseconds to minutes: time-resolved macromolecular crystallography at XFELs and synchrotrons
This review constitutes an overview of the current status of time-resolved crystallography performed at synchrotrons and XFELs on timescales ranging from femtoseconds to minutes. Methods, potential biases, instruments and examples are presented and compared with those for the cryo-trapping of reaction-intermediate states.
Open  access
access
 access
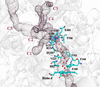
accessThe High-Pressure Freezing Laboratory for Macromolecular Crystallography (HPMX) at the ESRF allows the preparation of gas derivatives of macromolecular crystals suitable for X-ray diffraction data collection on macromolecular crystallography beamlines. Information obtained from pressurized crystals and/or gas-derivatized structures enables the improved understanding of specific issues in structural biology, such as the internal functional architecture of proteins, the interactions and reactivity of gases with macromolecules and functional structural changes including ligand-binding processes.
CCPEM
Open  access
access
 access
accessrelion_live.py and relion_analyse.py, two web-based tools to enhance cryo-EM data processing in RELION, are introduced, providing an interface for real-time feedback on data collection and simplified interpretation of metadata. Additionally, an analytical script for ice-quality estimation is provided, empowering researchers to make informed decisions to improve data quality and accessibility in cryo-EM.
research papers
Open  access
access
 access
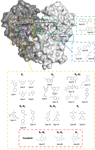
accessScytalidium thermophilum produces a catalase enzyme that is capable of oxidizing o-diphenolic and some p-diphenolic compounds in the absence of hydrogen peroxide. To better understand the role of the main channel in phenol oxidase activity, the gate residues Glu484 and Thr188 in the upper part of the main channel were investigated in a combined kinetic, spectroscopic and structural study.
Open  access
access
 access
accessStructural rearrangement of the Toscana virus nucleoprotein is induced by a single-chain camelid antibody.
PDB reference: Toscana virus nucleoprotein, 8rcq
Open  access
access
 access
accessX-ray crystallographic screening of SARS-CoV-2 3CL protease resulted in 29 fragment hits, including two isatin-based reversible covalent binders, and revealed a strong influence of the crystal form used for fragment soaking on the bound conformations of three additional reference fragments.
PDB references: SARS-CoV-2 main protease, complex with cpd-1, 7gre; complex with cpd-2, 7grf; complex with cpd-3, 7grg; complex with cpd-4, 7grh; complex with cpd-5, 7gri; complex with cpd-6, 7grj; complex with cpd-7, 7grk; complex with cpd-8, 7grl; complex with cpd-9, 7grm; complex with cpd-10, 7grn; complex with cpd-11, 7gro; complex with cpd-12, 7grp; complex with cpd-13, 7grq; complex with cpd-14, 7grr; complex with cpd-15, 7grs; complex with cpd-16, 7grt; complex with cpd-17, 7gru; complex with cpd-18, 7grv; complex with cpd-19, 7grw; complex with cpd-20, 7grx; complex with cpd-21, 7gry; complex with cpd-22, 7grz; complex with cpd-23, 7gs0; complex with cpd-24, 7gs1; complex with cpd-25, 7gs2; complex with cpd-26, 7gs3; complex with cpd-27, 7gs4; complex with cpd-28, 7gs5; complex with cpd-29, 7gs6
The crystal structure of a specific nanobody against NEIL1 was determined to 2.1 Å resolution. The structure was ultimately solved in an orthorhombic space group from ambiguous diffraction data.
PDB reference: glycosylase-specific nanobody, 8ftt

 journal menu
journal menu