Buy article online - an online subscription or single-article purchase is required to access this article.

The search for new tuberculostatics is an important issue due to the increasing resistance of
Mycobacterium tuberculosis to existing agents and the resulting spread of the pathogen. Heteroaryldithiocarbazic acid derivatives have shown potential tuberculostatic activity and investigations of the structural aspects of these compounds are thus of interest. Three new examples have been synthesized. The structure of methyl 2-[amino(pyridin-3-yl)methylidene]hydrazinecarbodithioate, C
8H
10N
4S
2, at 293 K has monoclinic (
P2
1/
n) symmetry. It is of interest with respect to antibacterial properties. The structure displays N—H

N and N—H

S hydrogen bonding. The structure of
N′-(pyrrolidine-1-carbonothioyl)picolinohydrazonamide, C
11H
15N
5S, at 100 K has monoclinic (
P2
1/
n) symmetry and is also of interest with respect to antibacterial properties. The structure displays N—H

S hydrogen bonding. The structure of (
Z)-methyl 2-[amino(pyridin-2-yl)methylidene]-1-methylhydrazinecarbodithioate, C
9H
13N
4S
2, has triclinic (
P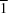
) symmetry. The structure displays N—H

S hydrogen bonding.
Supporting information
CCDC references: 1824482; 1824481; 1824480
Data collection: APEX2 (Bruker, 2009) for S1, S2; CrysAlis PRO (Rigaku OD, 2015) for S3. Cell refinement: SAINT-Plus (Bruker, 2009) for S1, S2; CrysAlis PRO (Rigaku OD, 2015) for S3. Data reduction: SAINT-Plus (Bruker, 2009) for S1, S2; CrysAlis PRO (Rigaku OD, 2015) for S3. For all structures, program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL2014 (Sheldrick, 2015); molecular graphics: PLATON (Spek, 2009) and Mercury (Macrae et al., 2008); software used to prepare material for publication: PLATON (Spek, 2009).
N'-(Pyrrolidine-1-carbonothioyl)picolinohydrazonamide (S1)
top
Crystal data top
| C11H15N5S | F(000) = 528 |
| Mr = 249.34 | Dx = 1.356 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54178 Å |
| a = 11.4343 (10) Å | Cell parameters from 9950 reflections |
| b = 7.7435 (7) Å | θ = 3.2–72.6° |
| c = 14.5125 (12) Å | µ = 2.24 mm−1 |
| β = 108.109 (2)° | T = 100 K |
| V = 1221.31 (18) Å3 | Needle, colourless |
| Z = 4 | 0.95 × 0.20 × 0.20 mm |
Data collection top
Bruker SMART APEXII CCD
diffractometer | 2371 reflections with I > 2σ(I) |
| ω scan | Rint = 0.032 |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2003) | θmax = 72.6°, θmin = 4.3° |
| Tmin = 0.698, Tmax = 1.000 | h = −13→14 |
| 14315 measured reflections | k = −9→9 |
| 2403 independent reflections | l = −17→17 |
Refinement top
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.031 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.082 | w = 1/[σ2(Fo2) + (0.0444P)2 + 0.5328P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 2403 reflections | Δρmax = 0.26 e Å−3 |
| 163 parameters | Δρmin = −0.25 e Å−3 |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.87777 (11) | 0.30274 (15) | 0.60567 (9) | 0.0188 (2) | |
| C4 | 0.76319 (11) | 0.37863 (15) | 0.79712 (8) | 0.0195 (3) | |
| C12 | 0.92173 (12) | 0.27575 (18) | 0.44829 (9) | 0.0246 (3) | |
| H12A | 0.9399 | 0.1531 | 0.4533 | 0.029* | |
| H12B | 0.9984 | 0.3400 | 0.4664 | 0.029* | |
| C13 | 0.84100 (13) | 0.32372 (19) | 0.34603 (9) | 0.0304 (3) | |
| H13A | 0.8909 | 0.3620 | 0.3067 | 0.036* | |
| H13B | 0.7914 | 0.2262 | 0.3145 | 0.036* | |
| C14 | 0.76038 (12) | 0.46970 (18) | 0.36197 (8) | 0.0251 (3) | |
| H14A | 0.8040 | 0.5789 | 0.3716 | 0.030* | |
| H14B | 0.6857 | 0.4803 | 0.3075 | 0.030* | |
| C15 | 0.73208 (11) | 0.41404 (17) | 0.45309 (8) | 0.0240 (3) | |
| H15A | 0.7159 | 0.5130 | 0.4882 | 0.029* | |
| H15B | 0.6618 | 0.3369 | 0.4378 | 0.029* | |
| C41 | 0.81293 (11) | 0.35985 (15) | 0.90393 (8) | 0.0192 (2) | |
| C42 | 0.74579 (11) | 0.40798 (16) | 0.96488 (9) | 0.0219 (3) | |
| H42 | 0.6654 | 0.4479 | 0.9398 | 0.026* | |
| C43 | 0.80202 (12) | 0.39482 (17) | 1.06441 (9) | 0.0245 (3) | |
| H43 | 0.7599 | 0.4263 | 1.1073 | 0.029* | |
| C44 | 0.92138 (12) | 0.33432 (18) | 1.09857 (9) | 0.0254 (3) | |
| H44 | 0.9617 | 0.3263 | 1.1648 | 0.030* | |
| C45 | 0.98018 (12) | 0.28551 (17) | 1.03183 (9) | 0.0251 (3) | |
| H45 | 1.0600 | 0.2429 | 1.0553 | 0.030* | |
| N2 | 0.79491 (9) | 0.35838 (14) | 0.64692 (7) | 0.0212 (2) | |
| N3 | 0.83570 (10) | 0.34140 (14) | 0.74619 (7) | 0.0202 (2) | |
| H3 | 0.9076 (16) | 0.305 (2) | 0.7743 (12) | 0.024* | |
| N5 | 0.64763 (10) | 0.42894 (15) | 0.75213 (8) | 0.0241 (2) | |
| H5A | 0.6312 (15) | 0.450 (2) | 0.6926 (13) | 0.029* | |
| H5B | 0.6050 (15) | 0.483 (2) | 0.7829 (11) | 0.029* | |
| N11 | 0.84526 (9) | 0.32455 (14) | 0.50907 (7) | 0.0209 (2) | |
| N46 | 0.92821 (10) | 0.29664 (14) | 0.93607 (8) | 0.0227 (2) | |
| S1 | 1.01620 (2) | 0.20426 (4) | 0.66657 (2) | 0.02183 (12) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0167 (6) | 0.0216 (6) | 0.0177 (6) | −0.0022 (4) | 0.0047 (4) | 0.0000 (4) |
| C4 | 0.0197 (6) | 0.0204 (6) | 0.0183 (6) | −0.0012 (4) | 0.0056 (5) | −0.0018 (4) |
| C12 | 0.0240 (6) | 0.0332 (7) | 0.0194 (6) | −0.0015 (5) | 0.0109 (5) | −0.0021 (5) |
| C13 | 0.0347 (7) | 0.0399 (8) | 0.0173 (6) | −0.0039 (6) | 0.0093 (5) | −0.0024 (5) |
| C14 | 0.0249 (6) | 0.0316 (7) | 0.0164 (6) | −0.0066 (5) | 0.0031 (5) | 0.0026 (5) |
| C15 | 0.0205 (6) | 0.0319 (7) | 0.0185 (6) | 0.0021 (5) | 0.0044 (5) | 0.0046 (5) |
| C41 | 0.0190 (6) | 0.0217 (6) | 0.0169 (6) | −0.0018 (4) | 0.0055 (5) | −0.0006 (4) |
| C42 | 0.0210 (6) | 0.0244 (6) | 0.0211 (6) | 0.0025 (5) | 0.0077 (5) | 0.0010 (5) |
| C43 | 0.0283 (7) | 0.0278 (6) | 0.0205 (6) | 0.0023 (5) | 0.0124 (5) | −0.0005 (5) |
| C44 | 0.0273 (7) | 0.0331 (7) | 0.0154 (6) | 0.0002 (5) | 0.0062 (5) | 0.0011 (5) |
| C45 | 0.0197 (6) | 0.0362 (7) | 0.0189 (6) | 0.0032 (5) | 0.0051 (5) | 0.0028 (5) |
| N2 | 0.0196 (5) | 0.0294 (6) | 0.0138 (5) | 0.0028 (4) | 0.0042 (4) | 0.0007 (4) |
| N3 | 0.0175 (5) | 0.0278 (5) | 0.0143 (5) | 0.0023 (4) | 0.0033 (4) | 0.0008 (4) |
| N5 | 0.0205 (5) | 0.0349 (6) | 0.0159 (5) | 0.0047 (4) | 0.0041 (4) | −0.0027 (4) |
| N11 | 0.0188 (5) | 0.0287 (5) | 0.0159 (5) | 0.0021 (4) | 0.0064 (4) | 0.0008 (4) |
| N46 | 0.0193 (5) | 0.0312 (6) | 0.0182 (5) | 0.0018 (4) | 0.0066 (4) | 0.0004 (4) |
| S1 | 0.01313 (17) | 0.0324 (2) | 0.01973 (17) | 0.00018 (10) | 0.00472 (12) | 0.00331 (11) |
Geometric parameters (Å, º) top
| C1—N2 | 1.3395 (16) | C14—C15 | 1.5183 (16) |
| C1—N11 | 1.3446 (15) | C15—N11 | 1.4702 (15) |
| C1—S1 | 1.7316 (12) | C41—N46 | 1.3462 (16) |
| C4—N3 | 1.3029 (16) | C41—C42 | 1.3904 (16) |
| C4—N5 | 1.3371 (16) | C42—C43 | 1.3903 (17) |
| C4—C41 | 1.4831 (16) | C43—C44 | 1.3810 (19) |
| C12—N11 | 1.4714 (15) | C44—C45 | 1.3921 (18) |
| C12—C13 | 1.5299 (18) | C45—N46 | 1.3336 (16) |
| C13—C14 | 1.521 (2) | N2—N3 | 1.3757 (13) |
| | | |
| N2—C1—N11 | 114.89 (10) | C42—C41—C4 | 122.04 (11) |
| N2—C1—S1 | 125.09 (9) | C43—C42—C41 | 118.22 (11) |
| N11—C1—S1 | 120.00 (9) | C44—C43—C42 | 118.96 (11) |
| N3—C4—N5 | 119.60 (11) | C43—C44—C45 | 118.65 (11) |
| N3—C4—C41 | 117.79 (11) | N46—C45—C44 | 123.53 (12) |
| N5—C4—C41 | 122.59 (11) | C1—N2—N3 | 112.37 (10) |
| N11—C12—C13 | 103.14 (10) | C4—N3—N2 | 120.30 (10) |
| C14—C13—C12 | 104.15 (10) | C1—N11—C15 | 123.09 (10) |
| C15—C14—C13 | 102.99 (10) | C1—N11—C12 | 124.72 (10) |
| N11—C15—C14 | 103.05 (10) | C15—N11—C12 | 112.01 (10) |
| N46—C41—C42 | 123.49 (11) | C45—N46—C41 | 117.11 (11) |
| N46—C41—C4 | 114.45 (10) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N5—H5B···S1i | 0.864 (17) | 2.456 (18) | 3.2895 (12) | 162.4 (14) |
| Symmetry code: (i) −x+3/2, y+1/2, −z+3/2. |
Methyl 2-[amino(pyridin-3-yl)methylidene]hydrazinecarbodithioate (S2)
top
Crystal data top
| C8H10N4S2 | F(000) = 472 |
| Mr = 226.32 | Dx = 1.443 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54178 Å |
| a = 7.2793 (9) Å | Cell parameters from 9829 reflections |
| b = 19.962 (2) Å | θ = 4.4–72.2° |
| c = 7.4299 (9) Å | µ = 4.36 mm−1 |
| β = 105.205 (3)° | T = 293 K |
| V = 1041.8 (2) Å3 | Needle, colourless |
| Z = 4 | 0.55 × 0.40 × 0.20 mm |
Data collection top
Bruker SMART APEXII CCD
diffractometer | 2043 reflections with I > 2σ(I) |
| ω scan | Rint = 0.042 |
Absorption correction: multi-scan
(SADABS; Sheldrick, 2003) | θmax = 72.3°, θmin = 4.4° |
| Tmin = 0.447, Tmax = 0.754 | h = −8→8 |
| 2048 measured reflections | k = 0→24 |
| 2048 independent reflections | l = 0→9 |
Refinement top
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.031 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.087 | w = 1/[σ2(Fo2) + (0.0506P)2 + 0.1572P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.12 | (Δ/σ)max < 0.001 |
| 2048 reflections | Δρmax = 0.27 e Å−3 |
| 138 parameters | Δρmin = −0.21 e Å−3 |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Refinement. Refined as a 2-component twin. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.7390 (2) | 0.13619 (8) | 0.1568 (2) | 0.0350 (3) | |
| S1 | 0.79198 (7) | 0.08918 (2) | 0.36434 (6) | 0.04270 (15) | |
| N2 | 0.7206 (2) | 0.20114 (7) | 0.18404 (18) | 0.0361 (3) | |
| H2 | 0.718 (3) | 0.2295 (11) | 0.099 (3) | 0.043* | |
| S2 | 0.70782 (7) | 0.10364 (2) | −0.05725 (6) | 0.04823 (16) | |
| N3 | 0.7569 (2) | 0.22656 (7) | 0.36600 (17) | 0.0362 (3) | |
| C4 | 0.6734 (2) | 0.28284 (7) | 0.3798 (2) | 0.0320 (3) | |
| N5 | 0.5633 (2) | 0.32035 (8) | 0.2418 (2) | 0.0403 (3) | |
| H5A | 0.523 (3) | 0.3024 (11) | 0.137 (3) | 0.048* | |
| H5B | 0.497 (3) | 0.3471 (11) | 0.277 (3) | 0.048* | |
| C11 | 0.8024 (4) | 0.00555 (9) | 0.2782 (3) | 0.0665 (6) | |
| H11A | 0.6801 | −0.0066 | 0.1992 | 0.100* | |
| H11B | 0.8368 | −0.0250 | 0.3813 | 0.100* | |
| H11C | 0.8958 | 0.0036 | 0.2080 | 0.100* | |
| C41 | 0.7107 (2) | 0.31150 (7) | 0.5706 (2) | 0.0314 (3) | |
| C42 | 0.7574 (2) | 0.37836 (8) | 0.6060 (2) | 0.0391 (3) | |
| H42 | 0.7608 | 0.4073 | 0.5089 | 0.047* | |
| C43 | 0.7989 (3) | 0.40134 (8) | 0.7878 (3) | 0.0445 (4) | |
| H43 | 0.8314 | 0.4459 | 0.8153 | 0.053* | |
| C44 | 0.7910 (3) | 0.35686 (9) | 0.9274 (2) | 0.0423 (4) | |
| H44 | 0.8234 | 0.3721 | 1.0500 | 0.051* | |
| C46 | 0.6995 (2) | 0.27098 (8) | 0.7191 (2) | 0.0360 (3) | |
| H46 | 0.6628 | 0.2265 | 0.6951 | 0.043* | |
| N46 | 0.7392 (2) | 0.29294 (7) | 0.89501 (18) | 0.0411 (3) | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0386 (7) | 0.0321 (7) | 0.0337 (8) | −0.0032 (6) | 0.0088 (6) | 0.0000 (6) |
| S1 | 0.0587 (3) | 0.0294 (2) | 0.0357 (2) | −0.00187 (16) | 0.00467 (18) | 0.00298 (14) |
| N2 | 0.0542 (8) | 0.0293 (6) | 0.0240 (6) | 0.0020 (5) | 0.0090 (5) | 0.0013 (5) |
| S2 | 0.0678 (3) | 0.0423 (2) | 0.0351 (2) | −0.01047 (19) | 0.0145 (2) | −0.01018 (16) |
| N3 | 0.0522 (7) | 0.0317 (6) | 0.0235 (6) | 0.0036 (5) | 0.0081 (5) | 0.0011 (5) |
| C4 | 0.0407 (7) | 0.0308 (7) | 0.0248 (7) | −0.0008 (6) | 0.0092 (6) | 0.0043 (5) |
| N5 | 0.0520 (8) | 0.0401 (8) | 0.0262 (6) | 0.0105 (6) | 0.0057 (6) | 0.0022 (5) |
| C11 | 0.0988 (17) | 0.0297 (9) | 0.0627 (13) | −0.0025 (10) | 0.0066 (12) | 0.0003 (9) |
| C41 | 0.0371 (7) | 0.0310 (7) | 0.0257 (7) | 0.0041 (6) | 0.0077 (5) | 0.0020 (6) |
| C42 | 0.0509 (9) | 0.0328 (8) | 0.0345 (8) | 0.0004 (6) | 0.0126 (7) | 0.0046 (6) |
| C43 | 0.0558 (10) | 0.0335 (8) | 0.0452 (10) | −0.0055 (7) | 0.0149 (8) | −0.0068 (7) |
| C44 | 0.0546 (9) | 0.0423 (9) | 0.0300 (8) | 0.0014 (7) | 0.0109 (7) | −0.0075 (6) |
| C46 | 0.0495 (8) | 0.0314 (7) | 0.0277 (7) | 0.0013 (6) | 0.0109 (6) | 0.0007 (6) |
| N46 | 0.0593 (8) | 0.0378 (7) | 0.0274 (6) | 0.0034 (6) | 0.0135 (6) | 0.0017 (5) |
Geometric parameters (Å, º) top
| C1—N2 | 1.324 (2) | C11—H11B | 0.9600 |
| C1—S2 | 1.6775 (15) | C11—H11C | 0.9600 |
| C1—S1 | 1.7592 (15) | C41—C42 | 1.385 (2) |
| S1—C11 | 1.797 (2) | C41—C46 | 1.388 (2) |
| N2—N3 | 1.4027 (17) | C42—C43 | 1.383 (2) |
| N2—H2 | 0.85 (2) | C42—H42 | 0.9300 |
| N3—C4 | 1.294 (2) | C43—C44 | 1.378 (3) |
| C4—N5 | 1.350 (2) | C43—H43 | 0.9300 |
| C4—C41 | 1.486 (2) | C44—N46 | 1.334 (2) |
| N5—H5A | 0.84 (2) | C44—H44 | 0.9300 |
| N5—H5B | 0.81 (2) | C46—N46 | 1.337 (2) |
| C11—H11A | 0.9600 | C46—H46 | 0.9300 |
| | | |
| N2—C1—S2 | 122.01 (12) | H11A—C11—H11C | 109.5 |
| N2—C1—S1 | 113.32 (11) | H11B—C11—H11C | 109.5 |
| S2—C1—S1 | 124.65 (9) | C42—C41—C46 | 118.26 (14) |
| C1—S1—C11 | 101.73 (9) | C42—C41—C4 | 121.76 (13) |
| C1—N2—N3 | 120.02 (13) | C46—C41—C4 | 119.99 (13) |
| C1—N2—H2 | 121.5 (14) | C43—C42—C41 | 119.00 (15) |
| N3—N2—H2 | 116.2 (14) | C43—C42—H42 | 120.5 |
| C4—N3—N2 | 114.62 (13) | C41—C42—H42 | 120.5 |
| N3—C4—N5 | 128.24 (15) | C44—C43—C42 | 118.69 (15) |
| N3—C4—C41 | 115.95 (13) | C44—C43—H43 | 120.7 |
| N5—C4—C41 | 115.74 (13) | C42—C43—H43 | 120.7 |
| C4—N5—H5A | 117.6 (15) | N46—C44—C43 | 123.09 (15) |
| C4—N5—H5B | 114.2 (16) | N46—C44—H44 | 118.5 |
| H5A—N5—H5B | 119 (2) | C43—C44—H44 | 118.5 |
| S1—C11—H11A | 109.5 | N46—C46—C41 | 122.94 (14) |
| S1—C11—H11B | 109.5 | N46—C46—H46 | 118.5 |
| H11A—C11—H11B | 109.5 | C41—C46—H46 | 118.5 |
| S1—C11—H11C | 109.5 | C44—N46—C46 | 117.93 (14) |
| | | |
| N2—C1—S1—C11 | 178.98 (14) | N5—C4—C41—C46 | 136.20 (15) |
| S2—C1—S1—C11 | 0.58 (15) | C46—C41—C42—C43 | 2.7 (2) |
| S2—C1—N2—N3 | −176.93 (11) | C4—C41—C42—C43 | −177.11 (15) |
| S1—C1—N2—N3 | 4.62 (19) | C41—C42—C43—C44 | −0.4 (3) |
| C1—N2—N3—C4 | −156.50 (15) | C42—C43—C44—N46 | −2.2 (3) |
| N2—N3—C4—N5 | −2.6 (2) | C42—C41—C46—N46 | −2.8 (2) |
| N2—N3—C4—C41 | −179.36 (12) | C4—C41—C46—N46 | 177.10 (14) |
| N3—C4—C41—C42 | 133.26 (16) | C43—C44—N46—C46 | 2.3 (3) |
| N5—C4—C41—C42 | −43.9 (2) | C41—C46—N46—C44 | 0.2 (2) |
| N3—C4—C41—C46 | −46.6 (2) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N5—H5A···N3i | 0.84 (2) | 2.47 (2) | 3.222 (2) | 150.2 (19) |
| N5—H5B···S2ii | 0.81 (2) | 2.88 (2) | 3.6405 (17) | 158.6 (19) |
| N2—H2···N46iii | 0.85 (2) | 2.01 (2) | 2.853 (2) | 174.1 (19) |
| Symmetry codes: (i) x−1/2, −y+1/2, z−1/2; (ii) x−1/2, −y+1/2, z+1/2; (iii) x, y, z−1. |
(
Z)-Methyl
2-[amino(pyridin-2-yl)methylidene]-1-methylhydrazinecarbodithioate (S3)
top
Crystal data top
| C9H12N4S2 | Z = 2 |
| Mr = 240.35 | F(000) = 252 |
| Triclinic, P1 | Dx = 1.362 Mg m−3 |
| a = 7.3230 (2) Å | Cu Kα radiation, λ = 1.54184 Å |
| b = 8.5983 (2) Å | Cell parameters from 21672 reflections |
| c = 10.0343 (2) Å | θ = 4.7–78.7° |
| α = 79.167 (2)° | µ = 3.91 mm−1 |
| β = 71.019 (2)° | T = 100 K |
| γ = 84.080 (2)° | Needle, colourless |
| V = 586.21 (3) Å3 | 0.50 × 0.40 × 0.15 mm |
Data collection top
XtaLAB Synergy, Dualflex, Pilatus 300K
diffractometer | 2417 independent reflections |
| Radiation source: micro-focus sealed X-ray tube | 2393 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.060 |
| ω scans | θmax = 79.0°, θmin = 4.7° |
Absorption correction: gaussian
(CrysAlis PRO; Rigaku OD, 2015) | h = −9→9 |
| Tmin = 0.176, Tmax = 1.000 | k = −10→10 |
| 24172 measured reflections | l = −12→12 |
Refinement top
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.035 | w = 1/[σ2(Fo2) + (0.0656P)2 + 0.309P]
where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.099 | (Δ/σ)max = 0.001 |
| S = 1.04 | Δρmax = 0.56 e Å−3 |
| 2417 reflections | Δρmin = −0.30 e Å−3 |
| 145 parameters | Extinction correction: SHELXL2014 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0147 (15) |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.31630 (19) | 0.09177 (17) | 0.72264 (15) | 0.0122 (3) | |
| S1 | 0.34397 (5) | 0.06346 (4) | 0.89411 (4) | 0.01573 (15) | |
| C2 | 0.3910 (2) | 0.27722 (18) | 0.49557 (16) | 0.0171 (3) | |
| H2A | 0.4059 | 0.1910 | 0.4439 | 0.026* | |
| H2B | 0.4892 | 0.3516 | 0.4443 | 0.026* | |
| H2C | 0.2658 | 0.3291 | 0.5056 | 0.026* | |
| N2 | 0.40933 (17) | 0.21624 (15) | 0.63707 (13) | 0.0135 (3) | |
| S2 | 0.18763 (5) | −0.02514 (4) | 0.67422 (4) | 0.01568 (15) | |
| N3 | 0.47965 (17) | 0.32513 (15) | 0.69812 (13) | 0.0144 (3) | |
| C4 | 0.6665 (2) | 0.30834 (17) | 0.67435 (15) | 0.0130 (3) | |
| N5 | 0.7889 (2) | 0.20436 (17) | 0.60358 (15) | 0.0204 (3) | |
| H5A | 0.752 (3) | 0.133 (3) | 0.569 (2) | 0.024* | |
| H5B | 0.899 (3) | 0.199 (3) | 0.603 (2) | 0.024* | |
| C11 | 0.2272 (2) | −0.11904 (19) | 0.97915 (17) | 0.0211 (3) | |
| H11A | 0.0966 | −0.1093 | 0.9764 | 0.032* | |
| H11B | 0.2264 | −0.1420 | 1.0767 | 0.032* | |
| H11C | 0.2960 | −0.2034 | 0.9299 | 0.032* | |
| C41 | 0.7511 (2) | 0.41881 (16) | 0.73531 (15) | 0.0124 (3) | |
| N42 | 0.94248 (18) | 0.40172 (16) | 0.70950 (15) | 0.0190 (3) | |
| C43 | 1.0231 (2) | 0.4971 (2) | 0.76351 (19) | 0.0229 (4) | |
| H43 | 1.1560 | 0.4864 | 0.7470 | 0.027* | |
| C44 | 0.9187 (2) | 0.6113 (2) | 0.84294 (18) | 0.0220 (3) | |
| H44 | 0.9803 | 0.6756 | 0.8784 | 0.026* | |
| C45 | 0.7210 (2) | 0.6273 (2) | 0.86809 (18) | 0.0219 (3) | |
| H45 | 0.6473 | 0.7030 | 0.9206 | 0.026* | |
| C46 | 0.6342 (2) | 0.52893 (19) | 0.81397 (16) | 0.0177 (3) | |
| H46 | 0.5015 | 0.5365 | 0.8299 | 0.021* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0091 (6) | 0.0145 (7) | 0.0155 (7) | 0.0010 (5) | −0.0050 (5) | −0.0071 (5) |
| S1 | 0.0181 (2) | 0.0174 (2) | 0.0156 (2) | −0.00557 (14) | −0.00900 (15) | −0.00283 (15) |
| C2 | 0.0170 (7) | 0.0195 (8) | 0.0162 (7) | −0.0027 (6) | −0.0067 (5) | −0.0029 (6) |
| N2 | 0.0131 (6) | 0.0136 (6) | 0.0166 (6) | −0.0027 (4) | −0.0067 (5) | −0.0046 (5) |
| S2 | 0.0159 (2) | 0.0162 (2) | 0.0195 (2) | −0.00468 (14) | −0.00815 (15) | −0.00725 (15) |
| N3 | 0.0127 (6) | 0.0137 (6) | 0.0200 (6) | −0.0032 (4) | −0.0065 (5) | −0.0065 (5) |
| C4 | 0.0146 (7) | 0.0122 (7) | 0.0135 (6) | −0.0019 (5) | −0.0060 (5) | −0.0020 (5) |
| N5 | 0.0130 (6) | 0.0253 (7) | 0.0298 (7) | 0.0014 (5) | −0.0091 (5) | −0.0181 (6) |
| C11 | 0.0237 (8) | 0.0178 (8) | 0.0230 (8) | −0.0058 (6) | −0.0106 (6) | 0.0021 (6) |
| C41 | 0.0139 (7) | 0.0108 (7) | 0.0138 (6) | −0.0030 (5) | −0.0059 (5) | −0.0010 (5) |
| N42 | 0.0140 (6) | 0.0181 (6) | 0.0277 (7) | −0.0016 (5) | −0.0070 (5) | −0.0094 (5) |
| C43 | 0.0152 (7) | 0.0233 (8) | 0.0354 (9) | −0.0039 (6) | −0.0101 (6) | −0.0116 (7) |
| C44 | 0.0227 (8) | 0.0208 (8) | 0.0286 (8) | −0.0064 (6) | −0.0114 (6) | −0.0100 (6) |
| C45 | 0.0222 (8) | 0.0211 (8) | 0.0268 (8) | −0.0008 (6) | −0.0081 (6) | −0.0136 (6) |
| C46 | 0.0147 (7) | 0.0203 (8) | 0.0207 (7) | −0.0011 (6) | −0.0060 (5) | −0.0088 (6) |
Geometric parameters (Å, º) top
| C1—N2 | 1.331 (2) | C11—H11A | 0.9600 |
| C1—S2 | 1.6767 (14) | C11—H11B | 0.9600 |
| C1—S1 | 1.7663 (14) | C11—H11C | 0.9600 |
| S1—C11 | 1.7947 (16) | C41—N42 | 1.3373 (19) |
| C2—N2 | 1.4626 (19) | C41—C46 | 1.388 (2) |
| C2—H2A | 0.9600 | N42—C43 | 1.338 (2) |
| C2—H2B | 0.9600 | C43—C44 | 1.388 (2) |
| C2—H2C | 0.9600 | C43—H43 | 0.9300 |
| N2—N3 | 1.4273 (17) | C44—C45 | 1.383 (2) |
| N3—C4 | 1.3073 (19) | C44—H44 | 0.9300 |
| C4—N5 | 1.3292 (19) | C45—C46 | 1.387 (2) |
| C4—C41 | 1.4991 (19) | C45—H45 | 0.9300 |
| N5—H5A | 0.86 (2) | C46—H46 | 0.9300 |
| N5—H5B | 0.80 (2) | | |
| | | |
| N2—C1—S2 | 124.23 (11) | S1—C11—H11B | 109.5 |
| N2—C1—S1 | 112.10 (10) | H11A—C11—H11B | 109.5 |
| S2—C1—S1 | 123.67 (9) | S1—C11—H11C | 109.5 |
| C1—S1—C11 | 102.69 (7) | H11A—C11—H11C | 109.5 |
| N2—C2—H2A | 109.5 | H11B—C11—H11C | 109.5 |
| N2—C2—H2B | 109.5 | N42—C41—C46 | 123.67 (13) |
| H2A—C2—H2B | 109.5 | N42—C41—C4 | 115.47 (12) |
| N2—C2—H2C | 109.5 | C46—C41—C4 | 120.85 (13) |
| H2A—C2—H2C | 109.5 | C41—N42—C43 | 117.22 (13) |
| H2B—C2—H2C | 109.5 | N42—C43—C44 | 123.42 (15) |
| C1—N2—N3 | 118.75 (12) | N42—C43—H43 | 118.3 |
| C1—N2—C2 | 123.12 (12) | C44—C43—H43 | 118.3 |
| N3—N2—C2 | 115.15 (12) | C45—C44—C43 | 118.43 (14) |
| C4—N3—N2 | 112.75 (11) | C45—C44—H44 | 120.8 |
| N3—C4—N5 | 127.57 (14) | C43—C44—H44 | 120.8 |
| N3—C4—C41 | 115.85 (12) | C44—C45—C46 | 119.14 (14) |
| N5—C4—C41 | 116.57 (13) | C44—C45—H45 | 120.4 |
| C4—N5—H5A | 122.6 (14) | C46—C45—H45 | 120.4 |
| C4—N5—H5B | 118.5 (15) | C45—C46—C41 | 118.12 (14) |
| H5A—N5—H5B | 118 (2) | C45—C46—H46 | 120.9 |
| S1—C11—H11A | 109.5 | C41—C46—H46 | 120.9 |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N5—H5B···S2i | 0.80 (2) | 2.88 (2) | 3.5431 (14) | 141.3 (19) |
| N5—H5A···S2ii | 0.86 (2) | 2.66 (2) | 3.3798 (14) | 141.5 (18) |
| Symmetry codes: (i) x+1, y, z; (ii) −x+1, −y, −z+1. |
The antimicrobial activity of compounds P1–P4 and
S1-S3 against Gram-positive bacteria
[MIC (MBC) (µg ml-1) values] top| Compound | S. aureus ATCC 25923 | S. aureus ATCC 6538 | S. aureus ATCC 43300 | S. epidermidis ATCC 12228 | M. luteus ATCC 10240 | B. subtilis ATCC 6633 | B. cereus ATCC 10876 | S. pyogenes ATCC 19615 | S. pneumoniae ATCC 49619 | S. mutans ATCC 25175 |
| P1 | >1000 | >1000 | | >1000 | 1000 | >1000 | | | | >1000 |
| P2 | 1000 (>1000) | 500 (>1000) | 500 (>1000) | >1000 (>1000) | >1000 (>1000) | 1000 (>1000) | >1000 (>1000) | 250 (>1000) | >1000 (>1000) | 250 (>1000) |
| P3 | 0.49 (7.9) | 3.9 (31.25) | 0.49 (0.49) | 0.49 (15.6) | 0.98 (7.8) | 0.49 (0.49) | 0.98 (3.9) | 15.6 (62.5) | 125 (250) | 7.8 (62.5) |
| P4 | 0.98 (1.95) | 1.95 (15.6) | 0.98 (15.6) | 0.98 (15.6) | 1.95 (1.95) | 1.95 (1.95) | 0.98 (0.98) | 31.25 (62.5) | 15.6 (62.5) | 7.8 (31.25) |
| S1 | 1.95 (125) | 7.8 (250) | 7.8 (250) | 7.8 (62.5) | 1.95 (3.9) | 7.8 (250) | 7.8 (250) | 15.6 (500) | 125 (250) | 125 (125) |
| S2 | 125 (500) | 500 (500) | 500 (500) | 250 (500) | 125 (250) | 250 (1000) | 250 (>1000) | 250 (500) | 500 (500) | 250 (250) |
| S3 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 |
| Vancomicin | 0.98 (7.81) | 0.49 (1.95) | 0.49 (1.95) | 0.98 (0.98) | 0.12 (0.12) | 0.24 (0.49) | 0.98 (15.6) | 0.24 (0.49) | 0.24 (0.49) | 0.98 (0.98) |
| Ciprofloxacin | 0.49 (0.49) | 0.24 (0.24) | 0.24 (0.24) | 0.49 (0.49) | 0.98 (1.95) | 0.03 (0.12) | 0.12 (0.12) | | | |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
[email protected] for assistance.
 N and N—H
N and N—H S hydrogen bonding. The structure of N′-(pyrrolidine-1-carbonothioyl)picolinohydrazonamide, C11H15N5S, at 100 K has monoclinic (P21/n) symmetry and is also of interest with respect to antibacterial properties. The structure displays N—H
S hydrogen bonding. The structure of N′-(pyrrolidine-1-carbonothioyl)picolinohydrazonamide, C11H15N5S, at 100 K has monoclinic (P21/n) symmetry and is also of interest with respect to antibacterial properties. The structure displays N—H S hydrogen bonding. The structure of (Z)-methyl 2-[amino(pyridin-2-yl)methylidene]-1-methylhydrazinecarbodithioate, C9H13N4S2, has triclinic (P
S hydrogen bonding. The structure of (Z)-methyl 2-[amino(pyridin-2-yl)methylidene]-1-methylhydrazinecarbodithioate, C9H13N4S2, has triclinic (P S hydrogen bonding.
S hydrogen bonding.
 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu











