Buy article online - an online subscription or single-article purchase is required to access this article.
The title compound, C
17H
16N
2O
3, is an antagonist for AMPA/kainate receptors. The molecule has its seven-membered oxadiazole ring in a boat conformation. Asymmetry of the two methoxy bond angles is evident, with (Me)O—C—C angles of 115.45 (12) and 124.78 (13)°, and 114.67 (12) and 125.31 (12)°. A centrosymmetric dimer involving the HN—CO moieties, with an N

O distance of 2.876 (2) Å, graph set
R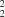
(8), is further linked into chains through methoxy C
sp3—H

N hydrogen bonds, with a C

N distance of 3.418 (2) Å.
Supporting information
CCDC reference: 174849
The title compound was obtained as described previously by De Sarro et
al. (1995). Suitable single crystals of (I) were obtained by
recrystallization from an ethanol solution.
The amine hydrogen H2 was located from a difference map and refined freely. The
remaining H atoms were located in idealized positions and allowed to ride on
their parent C atoms, with Uiso(H) = 1.2Ueq(C) and C—H =
0.93–0.97 Å.
Data collection: P3/V (Siemens, 1989); cell refinement: P3/V; data reduction: SHELXTL-Plus (Sheldrick, 1990); program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: XPW (Siemens, 1996); software used to prepare material for publication: PARST97 (Nardelli, 1995) and SHELXL97.
4,5-Dihydro-7,8-dimethoxy-1-phenyl-3
H-2,3-benzodiazepin-4-one
top
Crystal data top
| C17H16N2O3 | F(000) = 624 |
| Mr = 296.32 | Dx = 1.356 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.207 (2) Å | Cell parameters from 30 reflections |
| b = 10.312 (2) Å | θ = 6.4–13.8° |
| c = 17.236 (2) Å | µ = 0.09 mm−1 |
| β = 95.60 (1)° | T = 298 K |
| V = 1451.7 (5) Å3 | Irregular, colourless |
| Z = 4 | 0.26 × 0.24 × 0.12 mm |
Data collection top
Siemens P4
diffractometer | Rint = 0.008 |
| Radiation source: fine-focus sealed tube | θmax = 25.1°, θmin = 2.3° |
| Graphite monochromator | h = −1→9 |
| ω/2θ scans | k = −3→12 |
| 2903 measured reflections | l = −20→20 |
| 2567 independent reflections | 3 standard reflections every 197 reflections |
| 1734 reflections with I > 2σ(I) | intensity decay: none |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.032 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.080 | w = 1/[σ2(Fo2) + (0.0519P)2]
where P = (Fo2 + 2Fc2)/3 |
| S = 0.84 | (Δ/σ)max = 0.001 |
| 2567 reflections | Δρmax = 0.16 e Å−3 |
| 204 parameters | Δρmin = −0.15 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 1997), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0045 (9) |
Crystal data top
| C17H16N2O3 | V = 1451.7 (5) Å3 |
| Mr = 296.32 | Z = 4 |
| Monoclinic, P21/n | Mo Kα radiation |
| a = 8.207 (2) Å | µ = 0.09 mm−1 |
| b = 10.312 (2) Å | T = 298 K |
| c = 17.236 (2) Å | 0.26 × 0.24 × 0.12 mm |
| β = 95.60 (1)° | |
Data collection top
Siemens P4
diffractometer | Rint = 0.008 |
| 2903 measured reflections | 3 standard reflections every 197 reflections |
| 2567 independent reflections | intensity decay: none |
| 1734 reflections with I > 2σ(I) | |
Refinement top
| R[F2 > 2σ(F2)] = 0.032 | 0 restraints |
| wR(F2) = 0.080 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.84 | Δρmax = 0.16 e Å−3 |
| 2567 reflections | Δρmin = −0.15 e Å−3 |
| 204 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| C1 | 0.44184 (17) | 0.62315 (14) | 0.58707 (9) | 0.0392 (4) | |
| O1 | 0.56570 (12) | 0.64626 (10) | 0.55456 (7) | 0.0502 (3) | |
| N2 | 0.33046 (15) | 0.53575 (13) | 0.55707 (9) | 0.0431 (4) | |
| H2 | 0.360 (2) | 0.4848 (18) | 0.5208 (11) | 0.066 (6)* | |
| N3 | 0.19281 (14) | 0.48951 (12) | 0.58928 (8) | 0.0418 (3) | |
| C4 | 0.09083 (17) | 0.56974 (14) | 0.61556 (9) | 0.0349 (4) | |
| C5 | 0.10517 (16) | 0.71263 (13) | 0.61856 (8) | 0.0324 (3) | |
| C6 | 0.25402 (16) | 0.77126 (14) | 0.64210 (8) | 0.0329 (3) | |
| C7 | 0.40348 (16) | 0.68860 (15) | 0.66093 (9) | 0.0398 (4) | |
| H7A | 0.4951 | 0.7419 | 0.6814 | 0.048* | |
| H7B | 0.383 | 0.6243 | 0.6999 | 0.048* | |
| C8 | 0.26573 (16) | 0.90642 (14) | 0.64534 (8) | 0.0360 (4) | |
| H8 | 0.3653 | 0.9453 | 0.662 | 0.043* | |
| C9 | 0.13049 (17) | 0.98247 (13) | 0.62402 (8) | 0.0344 (4) | |
| C10 | −0.01944 (15) | 0.92377 (14) | 0.59857 (8) | 0.0318 (3) | |
| C11 | −0.03263 (16) | 0.79086 (14) | 0.59738 (8) | 0.0332 (3) | |
| H11 | −0.1333 | 0.7523 | 0.5825 | 0.04* | |
| C12 | −0.05541 (16) | 0.50662 (13) | 0.64471 (9) | 0.0361 (4) | |
| C13 | −0.11398 (19) | 0.38906 (15) | 0.61385 (10) | 0.0489 (4) | |
| H13 | −0.0617 | 0.3488 | 0.5748 | 0.059* | |
| C14 | −0.2495 (2) | 0.33216 (17) | 0.64108 (12) | 0.0632 (5) | |
| H14 | −0.2881 | 0.2537 | 0.62 | 0.076* | |
| C15 | −0.3284 (2) | 0.38962 (17) | 0.69880 (11) | 0.0574 (5) | |
| H15 | −0.4208 | 0.3511 | 0.7161 | 0.069* | |
| C16 | −0.26981 (19) | 0.50438 (16) | 0.73070 (10) | 0.0478 (4) | |
| H16 | −0.3209 | 0.543 | 0.7707 | 0.057* | |
| C17 | −0.13526 (17) | 0.56232 (15) | 0.70350 (9) | 0.0401 (4) | |
| H17 | −0.0972 | 0.6406 | 0.7251 | 0.048* | |
| O2 | 0.12966 (12) | 1.11411 (10) | 0.62487 (7) | 0.0481 (3) | |
| C18 | 0.27701 (19) | 1.17974 (16) | 0.65132 (10) | 0.0524 (4) | |
| H18A | 0.259 | 1.2717 | 0.6488 | 0.079* | |
| H18B | 0.3611 | 1.1569 | 0.6188 | 0.079* | |
| H18C | 0.3105 | 1.155 | 0.7042 | 0.079* | |
| O3 | −0.14385 (11) | 1.00856 (9) | 0.57731 (6) | 0.0418 (3) | |
| C19 | −0.30176 (18) | 0.95579 (17) | 0.55732 (11) | 0.0521 (5) | |
| H19A | −0.3779 | 1.0247 | 0.5436 | 0.078* | |
| H19B | −0.336 | 0.9088 | 0.601 | 0.078* | |
| H19C | −0.2985 | 0.8981 | 0.5138 | 0.078* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| C1 | 0.0325 (8) | 0.0317 (9) | 0.0543 (10) | 0.0061 (7) | 0.0089 (7) | 0.0012 (7) |
| O1 | 0.0402 (6) | 0.0439 (7) | 0.0694 (8) | −0.0028 (5) | 0.0198 (5) | −0.0096 (6) |
| N2 | 0.0391 (7) | 0.0366 (8) | 0.0565 (9) | −0.0011 (6) | 0.0189 (6) | −0.0107 (7) |
| N3 | 0.0386 (7) | 0.0325 (7) | 0.0563 (9) | 0.0015 (6) | 0.0153 (6) | −0.0026 (6) |
| C4 | 0.0346 (8) | 0.0303 (8) | 0.0400 (9) | 0.0016 (7) | 0.0048 (6) | −0.0014 (7) |
| C5 | 0.0327 (7) | 0.0285 (8) | 0.0371 (8) | 0.0009 (6) | 0.0089 (6) | −0.0010 (7) |
| C6 | 0.0325 (8) | 0.0320 (8) | 0.0350 (8) | 0.0035 (6) | 0.0071 (6) | −0.0019 (7) |
| C7 | 0.0339 (8) | 0.0373 (9) | 0.0484 (10) | 0.0050 (7) | 0.0044 (7) | −0.0028 (7) |
| C8 | 0.0303 (8) | 0.0355 (9) | 0.0423 (9) | −0.0029 (6) | 0.0045 (6) | −0.0052 (7) |
| C9 | 0.0374 (8) | 0.0266 (8) | 0.0399 (9) | 0.0002 (6) | 0.0076 (7) | −0.0006 (7) |
| C10 | 0.0296 (7) | 0.0318 (8) | 0.0346 (8) | 0.0035 (6) | 0.0061 (6) | 0.0016 (6) |
| C11 | 0.0278 (7) | 0.0331 (8) | 0.0391 (8) | −0.0023 (6) | 0.0055 (6) | −0.0014 (7) |
| C12 | 0.0346 (8) | 0.0285 (8) | 0.0451 (9) | 0.0018 (6) | 0.0039 (7) | 0.0052 (7) |
| C13 | 0.0502 (9) | 0.0319 (9) | 0.0657 (11) | −0.0016 (8) | 0.0117 (8) | −0.0032 (8) |
| C14 | 0.0595 (11) | 0.0369 (10) | 0.0941 (15) | −0.0148 (9) | 0.0118 (10) | 0.0001 (10) |
| C15 | 0.0476 (10) | 0.0467 (11) | 0.0797 (14) | −0.0067 (9) | 0.0158 (9) | 0.0193 (10) |
| C16 | 0.0443 (9) | 0.0513 (11) | 0.0494 (11) | 0.0034 (8) | 0.0126 (8) | 0.0121 (8) |
| C17 | 0.0385 (8) | 0.0376 (9) | 0.0444 (9) | 0.0007 (7) | 0.0049 (7) | 0.0007 (7) |
| O2 | 0.0446 (6) | 0.0258 (6) | 0.0725 (8) | −0.0016 (5) | −0.0011 (5) | −0.0018 (5) |
| C18 | 0.0522 (10) | 0.0334 (9) | 0.0703 (12) | −0.0088 (8) | −0.0006 (8) | −0.0044 (9) |
| O3 | 0.0332 (5) | 0.0333 (6) | 0.0583 (7) | 0.0049 (5) | 0.0008 (5) | 0.0039 (5) |
| C19 | 0.0348 (8) | 0.0514 (11) | 0.0678 (12) | 0.0046 (8) | −0.0058 (8) | −0.0032 (9) |
Geometric parameters (Å, º) top
| C1—O1 | 1.231 (2) | C13—C14 | 1.380 (2) |
| C1—N2 | 1.350 (2) | C14—C15 | 1.373 (2) |
| C1—C7 | 1.501 (2) | C15—C16 | 1.372 (2) |
| N2—N3 | 1.391 (2) | C16—C17 | 1.377 (2) |
| N3—C4 | 1.290 (2) | N2—H2 | 0.87 (2) |
| C4—C5 | 1.479 (2) | C7—H7A | 0.97 |
| C4—C12 | 1.495 (2) | C7—H7B | 0.97 |
| C5—C6 | 1.388 (2) | C8—H8 | 0.93 |
| C5—C11 | 1.4080 (19) | C11—H11 | 0.93 |
| C6—C8 | 1.398 (2) | C13—H13 | 0.93 |
| C6—C7 | 1.503 (2) | C14—H14 | 0.93 |
| C8—C9 | 1.3793 (19) | C15—H15 | 0.93 |
| O2—C9 | 1.358 (2) | C16—H16 | 0.93 |
| O3—C10 | 1.367 (2) | C17—H17 | 0.93 |
| O2—C18 | 1.421 (2) | C18—H18A | 0.96 |
| O3—C19 | 1.417 (2) | C18—H18B | 0.96 |
| C9—C10 | 1.4027 (19) | C18—H18C | 0.96 |
| C10—C11 | 1.375 (2) | C19—H19A | 0.96 |
| C12—C17 | 1.384 (2) | C19—H19B | 0.96 |
| C12—C13 | 1.391 (2) | C19—H19C | 0.96 |
| | | |
| O1—C1—N2 | 120.9 (1) | C5—C11—H11 | 119.8 |
| O1—C1—C7 | 123.4 (1) | C17—C12—C13 | 118.11 (14) |
| N2—C1—C7 | 115.8 (1) | C17—C12—C4 | 121.23 (13) |
| C1—N2—N3 | 128.5 (1) | C13—C12—C4 | 120.66 (14) |
| C1—N2—H2 | 117 (1) | C14—C13—C12 | 120.04 (16) |
| N3—N2—H2 | 112 (1) | C14—C13—H13 | 120 |
| N2—N3—C4 | 120.0 (1) | C12—C13—H13 | 120 |
| N3—C4—C5 | 126.8 (1) | C15—C14—C13 | 121.04 (17) |
| N3—C4—C12 | 114.1 (1) | C15—C14—H14 | 119.5 |
| C5—C4—C12 | 119.1 (1) | C13—C14—H14 | 119.5 |
| C6—C5—C11 | 119.22 (13) | C16—C15—C14 | 119.39 (16) |
| C4—C5—C6 | 120.6 (1) | C16—C15—H15 | 120.3 |
| C11—C5—C4 | 120.18 (13) | C14—C15—H15 | 120.3 |
| C5—C6—C8 | 120.08 (13) | C15—C16—C17 | 119.95 (16) |
| C5—C6—C7 | 119.5 (1) | C15—C16—H16 | 120 |
| C8—C6—C7 | 120.36 (13) | C17—C16—H16 | 120 |
| C1—C7—C6 | 107.8 (1) | C16—C17—C12 | 121.45 (15) |
| C1—C7—H7A | 110.1 | C16—C17—H17 | 119.3 |
| C6—C7—H7A | 110.1 | C12—C17—H17 | 119.3 |
| C1—C7—H7B | 110.1 | C9—O2—C18 | 118.3 (1) |
| C6—C7—H7B | 110.1 | O2—C18—H18A | 109.5 |
| H7A—C7—H7B | 108.5 | O2—C18—H18B | 109.5 |
| C9—C8—C6 | 120.39 (13) | H18A—C18—H18B | 109.5 |
| C9—C8—H8 | 119.8 | O2—C18—H18C | 109.5 |
| C6—C8—H8 | 119.8 | H18A—C18—H18C | 109.5 |
| O2—C9—C8 | 124.78 (13) | H18B—C18—H18C | 109.5 |
| O2—C9—C10 | 115.45 (12) | C10—O3—C19 | 117.5 (1) |
| C8—C9—C10 | 119.77 (13) | O3—C19—H19A | 109.5 |
| O3—C10—C11 | 125.31 (12) | O3—C19—H19B | 109.5 |
| O3—C10—C9 | 114.67 (12) | H19A—C19—H19B | 109.5 |
| C11—C10—C9 | 120.01 (13) | O3—C19—H19C | 109.5 |
| C10—C11—C5 | 120.48 (13) | H19A—C19—H19C | 109.5 |
| C10—C11—H11 | 119.8 | H19B—C19—H19C | 109.5 |
| | | |
| O1—C1—N2—N3 | 173.64 (14) | O2—C9—C10—C11 | −178.38 (13) |
| C7—C1—N2—N3 | −6.6 (2) | C8—C9—C10—C11 | 2.2 (2) |
| C1—N2—N3—C4 | 50.0 (2) | O3—C10—C11—C5 | 178.23 (13) |
| N2—N3—C4—C5 | −2.5 (2) | C9—C10—C11—C5 | −2.6 (2) |
| N2—N3—C4—C12 | 177.05 (13) | C6—C5—C11—C10 | 1.2 (2) |
| N3—C4—C5—C6 | −42.8 (2) | C4—C5—C11—C10 | −178.71 (13) |
| C12—C4—C5—C6 | 137.75 (14) | N3—C4—C12—C17 | 150.78 (14) |
| N3—C4—C5—C11 | 137.15 (16) | C5—C4—C12—C17 | −29.7 (2) |
| C12—C4—C5—C11 | −42.3 (2) | N3—C4—C12—C13 | −28.5 (2) |
| C11—C5—C6—C8 | 0.6 (2) | C5—C4—C12—C13 | 151.02 (15) |
| C4—C5—C6—C8 | −179.45 (14) | C17—C12—C13—C14 | 1.0 (2) |
| C11—C5—C6—C7 | −176.94 (12) | C4—C12—C13—C14 | −179.72 (15) |
| C4—C5—C6—C7 | 3.0 (2) | C12—C13—C14—C15 | −0.2 (3) |
| O1—C1—C7—C6 | 113.45 (16) | C13—C14—C15—C16 | −1.1 (3) |
| N2—C1—C7—C6 | −66.28 (17) | C14—C15—C16—C17 | 1.5 (3) |
| C5—C6—C7—C1 | 65.53 (17) | C15—C16—C17—C12 | −0.7 (2) |
| C8—C6—C7—C1 | −112.03 (15) | C13—C12—C17—C16 | −0.5 (2) |
| C5—C6—C8—C9 | −1.1 (2) | C4—C12—C17—C16 | −179.83 (13) |
| C7—C6—C8—C9 | 176.48 (13) | C8—C9—O2—C18 | −1.6 (2) |
| C6—C8—C9—O2 | −179.73 (14) | C10—C9—O2—C18 | 178.95 (13) |
| C6—C8—C9—C10 | −0.3 (2) | C11—C10—O3—C19 | 4.4 (2) |
| O2—C9—C10—O3 | 0.88 (19) | C9—C10—O3—C19 | −174.81 (13) |
| C8—C9—C10—O3 | −178.59 (12) | | |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O1i | 0.87 (2) | 2.01 (2) | 2.876 (2) | 174 (2) |
| C18—H18A···N3ii | 0.96 | 2.51 | 3.418 (2) | 158 |
| C13—H13···N3 | 0.93 | 2.54 | 2.793 (2) | 96 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y+1, z. |
Experimental details
| Crystal data |
| Chemical formula | C17H16N2O3 |
| Mr | 296.32 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 298 |
| a, b, c (Å) | 8.207 (2), 10.312 (2), 17.236 (2) |
| β (°) | 95.60 (1) |
| V (Å3) | 1451.7 (5) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.09 |
| Crystal size (mm) | 0.26 × 0.24 × 0.12 |
| |
| Data collection |
| Diffractometer | Siemens P4
diffractometer |
| Absorption correction | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 2903, 2567, 1734 |
| Rint | 0.008 |
| (sin θ/λ)max (Å−1) | 0.597 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.032, 0.080, 0.84 |
| No. of reflections | 2567 |
| No. of parameters | 204 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.16, −0.15 |
Selected geometric parameters (Å, º) top| C1—O1 | 1.231 (2) | C5—C6 | 1.388 (2) |
| C1—N2 | 1.350 (2) | C6—C7 | 1.503 (2) |
| C1—C7 | 1.501 (2) | O2—C9 | 1.358 (2) |
| N2—N3 | 1.391 (2) | O3—C10 | 1.367 (2) |
| N3—C4 | 1.290 (2) | O2—C18 | 1.421 (2) |
| C4—C5 | 1.479 (2) | O3—C19 | 1.417 (2) |
| | | |
| O1—C1—N2 | 120.9 (1) | C4—C5—C6 | 120.6 (1) |
| O1—C1—C7 | 123.4 (1) | C5—C6—C7 | 119.5 (1) |
| C1—N2—N3 | 128.5 (1) | C9—O2—C18 | 118.3 (1) |
| N2—N3—C4 | 120.0 (1) | C10—O3—C19 | 117.5 (1) |
| N3—C4—C5 | 126.8 (1) | | |
| | | |
| N2—C1—C7—C6 | −66.28 (17) | C8—C9—O2—C18 | −1.6 (2) |
| C5—C4—C12—C17 | −29.7 (2) | C11—C10—O3—C19 | 4.4 (2) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O1i | 0.87 (2) | 2.01 (2) | 2.876 (2) | 174 (2) |
| C18—H18A···N3ii | 0.96 | 2.51 | 3.418 (2) | 158 |
| C13—H13···N3 | 0.93 | 2.54 | 2.793 (2) | 96 |
| Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y+1, z. |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
[email protected] for assistance.
 O distance of 2.876 (2) Å, graph set R
O distance of 2.876 (2) Å, graph set R N hydrogen bonds, with a C
N hydrogen bonds, with a C N distance of 3.418 (2) Å.
N distance of 3.418 (2) Å.
 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
 journal menu
journal menu








![[Figure 1]](https://journals.iucr.org/c/issues/2001/10/00/gg1067/gg1067fig1thm.gif)
![[Figure 2]](https://journals.iucr.org/c/issues/2001/10/00/gg1067/gg1067fig2thm.gif)




In previous publications (De Sarro et al., 1995; Chimirri et al., 1997, 1998), we reported on chemical and biological studies of some 2,3-benzodiazepine derivatives which have been shown to behave as anticonvulsants, acting as non-competitive antagonists against AMPA/kainate receptors. This paper describes the crystal structure analysis of 4,5-dihydro-7,8-dimethoxy-1-phenyl-3H-2,3-benzodiazepin-4-one, (I), one of the most active compounds of the series, with the aim of comparing the molecular geometry with the analogous 2,3-benzodiazepines reported in the literature (Anderson et al., 1996). The results of this study will be used for the study of structure-activity relationships, in order to understand better the structural characteristics necessary for AMPA/kainate receptor antagonists. \sch
Although there are several crystal structure reports on benzodiazepines, the 1,2- or 2,3-derivatives are relatively rare. In (I) (Fig. 1), the bond lengths and angles of the seven-membered ring are in good agreement with the corresponding values reported for the parent compound 7,11-dihydro-9,10-dimethoxy-3,11b-diphenyl[1,2,4]oxadiazolo[5,4-a][2,3]benzodiazepin-6(5H)-one, (II) (Bruno et al., 1999), and for the rare analogous 2,3-benzodiazepine reported in the Cambridge Structural Database (CSD; April 2001 release, version 5.21; Allen & Kennard, 1993).
In the diazepinone fragment, the bond lengths agree reasonably well with the system of localized single and double bonds (Table 1). Slight deviations from the expected values may be explained by the intra- and intermolecular hydrogen bonds involving this fragment. Atom N3 is involved in an intramolecular contact with the H atom on C13, and this contact is critical for the orientation of the phenyl ring bonded to C4. A strong intermolecular hydrogen bond involves a pair of molecules related by the inversion centre through the HN—CO groups, with graph set R22(8) (Bernstein et al., 1995). This centrosymmetric dimer is connected to adjacent units by intermolecular methoxy C18—H18A···N3 hydrogen bonds, graph set R44(24), along the b axis. Both intra- and intermolecular contacts and interactions are responsible for the molecular packing observed in the solid state.
The seven-membered ring has a boat conformation [ϕ2 = 53.9 (1), ϕ3 = -102.7 (4) and θ = 76.6 (1)°, and Q = 0.892 (2) Å; Cremer & Pople, 1975). As in compound (II) (Bruno et al., 1999), and in the organic structures in the CSD containing the fragment depicted in Fig. 2, in (I) a large asymmetry in the dimethoxy ring angles is noticeable: O2—C9—C10 = 115.5 (1), O2—C9—C8 = 124.8 (1), O3—C10—C9 = 114.7 (1) and O3—C10—C11 = 125.3 (1)°. Although the planarity of the methoxy groups with the phenyl ring was noted in previous work, these authors attributed such an orientation either to the `presumable' steric interaction between the methyl and phenyl H atoms (Salem et al., 1988; Sharma et al., 1997; Dijksma et al., 1998; Kumar et al., 1998) or to the correct balance between no (non-bonding orbital) and the π delocalization of the oxygen lone pairs and the cited H···H interaction (Bock et al., 1995). The latter explanation was also proposed in a statistical analysis of crystal structure data for mono-substituted methoxyphenyl compounds (Hummel et al., 1988). The significant asymmetry in the above methoxyphenyl angles [mean 10.0 (2)°] also causes a close contact of 2.557 (2) Å between the two methoxy O atoms, which is smaller than their van der Waals radii sum (2.80 Å; ref?).
A search was undertaken of the CSD database and 478 organic compounds (with R < 0.081) were located containing the fragment shown in Fig. 2, which gives the mean values for the significant structural parameters. We deduce that the planarity of the methoxy group with respect to the phenyl ring is determined by conjugation effects. In order to rationalize this problem still further, we have in progress research into statistical analysis on methoxy-phenyl substituted compounds, and are carrying out semi-empirical and ab initio calculations on this fragment model. This research will be the subject of a forthcoming publication (Bruno & Nicoló, 2001).