Buy article online - an online subscription or single-article purchase is required to access this article.
Bis(4,5-diamino-1,2,4-triazol-3-yl)methane monohydrate (BDATZM·H
2O or C
5H
10N
10·H
2O) was synthesized and its crystal structure characterized by single-crystal X-ray diffraction; it belongs to the space group
P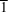
(triclinic) with
Z = 2. The structure of BDATZM·H
2O can be described as a two-dimensional ladder plane with extensive hydrogen bonding and no disorder. The thermal behaviour was studied under non-isothermal conditions by differential scanning calorimetry (DSC) and thermogravimetric/differential thermogravimetric (TG/DTG) methods. The detonation velocity (
D) and detonation pressure (
P) of BDATZM were estimated using the nitrogen equivalent equation according to the experimental density. A comparison between BDATZM·H
2O and bis(5-amino-1,2,4-triazol-3-yl)methane (BATZM) was made to determine the effect of the amino group; the results suggest that the amino group increases the hydrophilicity, space utilization and energy, and decreases the thermal stability and symmetry of the resulting compound.
Supporting information
CCDC reference: 1060501
Data collection: APEX2 (Bruker, 2009); cell refinement: SAINT (Bruker, 2009); data reduction: SAINT (Bruker, 2009); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008); program(s) used to refine structure: SHELXL2018 (Sheldrick, 2015); molecular graphics: SHELXTL (Sheldrick, 2008); software used to prepare material for publication: SHELXTL (Sheldrick, 2008).
Bis(4,5-diamino-1,2,4-triazol-3-yl)methane monohydrate
top
Crystal data top
| C5H10N10·H2O | Z = 2 |
| Mr = 228.25 | F(000) = 240 |
| Triclinic, P1 | Dx = 1.580 Mg m−3 |
| a = 5.4364 (10) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 5.6202 (11) Å | Cell parameters from 1383 reflections |
| c = 15.844 (3) Å | θ = 2.6–28.0° |
| α = 83.623 (3)° | µ = 0.12 mm−1 |
| β = 87.617 (3)° | T = 296 K |
| γ = 86.146 (3)° | Rodlike, colourless |
| V = 479.71 (16) Å3 | 0.34 × 0.28 × 0.15 mm |
Data collection top
Bruker APEXII CCD
diffractometer | 1522 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.011 |
Absorption correction: multi-scan
(SADABS; Bruker, 2009) | θmax = 25.1°, θmin = 2.6° |
| Tmin = 0.960, Tmax = 0.982 | h = −5→6 |
| 2421 measured reflections | k = −6→6 |
| 1698 independent reflections | l = −18→15 |
Refinement top
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.035 | All H-atom parameters refined |
| wR(F2) = 0.092 | w = 1/[σ2(Fo2) + (0.0439P)2 + 0.179P]
where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 1698 reflections | Δρmax = 0.17 e Å−3 |
| 194 parameters | Δρmin = −0.15 e Å−3 |
| 0 restraints | Extinction correction: SHELXL2018 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.107 (8) |
Special details top
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell esds are taken
into account individually in the estimation of esds in distances, angles
and torsion angles; correlations between esds in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| N3 | 0.4877 (2) | 0.6949 (2) | 0.37479 (8) | 0.0284 (3) | |
| N8 | 0.4006 (2) | 0.4532 (2) | 0.12692 (8) | 0.0292 (3) | |
| N1 | 0.1762 (3) | 0.4668 (3) | 0.40038 (9) | 0.0411 (4) | |
| N7 | 0.7210 (3) | 0.2065 (2) | 0.15392 (9) | 0.0388 (4) | |
| N10 | 0.2307 (3) | 0.6532 (3) | 0.12384 (11) | 0.0408 (4) | |
| N5 | 0.6456 (3) | 0.8754 (3) | 0.38523 (11) | 0.0448 (4) | |
| N6 | 0.5896 (3) | 0.1227 (2) | 0.08966 (9) | 0.0383 (4) | |
| N2 | 0.3076 (3) | 0.4056 (3) | 0.32649 (8) | 0.0390 (4) | |
| C5 | 0.3981 (3) | 0.2755 (3) | 0.07553 (9) | 0.0308 (4) | |
| C2 | 0.4888 (3) | 0.5453 (3) | 0.31288 (9) | 0.0293 (4) | |
| N9 | 0.2256 (3) | 0.2702 (3) | 0.01670 (10) | 0.0425 (4) | |
| C4 | 0.6046 (3) | 0.4023 (3) | 0.17430 (9) | 0.0305 (4) | |
| N4 | 0.2268 (3) | 0.7560 (3) | 0.49553 (10) | 0.0484 (4) | |
| C1 | 0.2906 (3) | 0.6410 (3) | 0.42740 (9) | 0.0318 (4) | |
| C3 | 0.6737 (3) | 0.5516 (3) | 0.24052 (10) | 0.0339 (4) | |
| O1 | 0.1062 (3) | −0.0044 (2) | 0.26006 (9) | 0.0502 (4) | |
| H3A | 0.832 (3) | 0.492 (3) | 0.2612 (10) | 0.032 (4)* | |
| H3B | 0.685 (3) | 0.718 (4) | 0.2154 (12) | 0.045 (5)* | |
| H4A | 0.103 (4) | 0.708 (4) | 0.5279 (13) | 0.050 (6)* | |
| H9A | 0.248 (4) | 0.148 (4) | −0.0139 (13) | 0.051 (6)* | |
| H10B | 0.187 (4) | 0.683 (4) | 0.1761 (16) | 0.074 (7)* | |
| H4B | 0.308 (4) | 0.875 (4) | 0.5077 (14) | 0.063 (6)* | |
| H1A | 0.171 (5) | 0.122 (5) | 0.2784 (15) | 0.075 (7)* | |
| H1B | −0.012 (5) | 0.059 (4) | 0.2209 (16) | 0.079 (8)* | |
| H9B | 0.079 (5) | 0.327 (4) | 0.0279 (15) | 0.070 (7)* | |
| H10A | 0.316 (5) | 0.782 (5) | 0.1007 (16) | 0.081 (8)* | |
| H5B | 0.797 (5) | 0.819 (4) | 0.3807 (15) | 0.075 (8)* | |
| H5A | 0.622 (6) | 0.995 (6) | 0.344 (2) | 0.122 (13)* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| N3 | 0.0293 (6) | 0.0291 (7) | 0.0289 (6) | −0.0081 (5) | 0.0001 (5) | −0.0088 (5) |
| N8 | 0.0327 (7) | 0.0264 (6) | 0.0292 (7) | −0.0026 (5) | 0.0021 (5) | −0.0070 (5) |
| N1 | 0.0460 (8) | 0.0476 (8) | 0.0336 (7) | −0.0205 (7) | 0.0095 (6) | −0.0153 (6) |
| N7 | 0.0399 (8) | 0.0411 (8) | 0.0379 (8) | 0.0022 (6) | −0.0061 (6) | −0.0166 (6) |
| N10 | 0.0445 (9) | 0.0331 (8) | 0.0450 (9) | 0.0048 (7) | −0.0003 (7) | −0.0094 (7) |
| N5 | 0.0426 (9) | 0.0477 (9) | 0.0502 (10) | −0.0233 (7) | 0.0070 (7) | −0.0232 (8) |
| N6 | 0.0431 (8) | 0.0372 (8) | 0.0372 (8) | 0.0018 (6) | −0.0056 (6) | −0.0163 (6) |
| N2 | 0.0453 (8) | 0.0417 (8) | 0.0336 (7) | −0.0189 (6) | 0.0074 (6) | −0.0142 (6) |
| C5 | 0.0361 (8) | 0.0307 (8) | 0.0262 (8) | −0.0049 (7) | 0.0020 (6) | −0.0059 (6) |
| C2 | 0.0332 (8) | 0.0278 (7) | 0.0284 (8) | −0.0059 (6) | −0.0022 (6) | −0.0070 (6) |
| N9 | 0.0435 (9) | 0.0482 (9) | 0.0385 (8) | −0.0007 (7) | −0.0063 (7) | −0.0162 (7) |
| C4 | 0.0317 (8) | 0.0318 (8) | 0.0289 (8) | −0.0053 (6) | 0.0030 (6) | −0.0070 (6) |
| N4 | 0.0509 (9) | 0.0557 (10) | 0.0444 (9) | −0.0241 (8) | 0.0177 (7) | −0.0265 (7) |
| C1 | 0.0330 (8) | 0.0347 (8) | 0.0289 (8) | −0.0075 (7) | 0.0016 (6) | −0.0066 (6) |
| C3 | 0.0335 (9) | 0.0372 (9) | 0.0336 (8) | −0.0089 (7) | 0.0019 (7) | −0.0118 (7) |
| O1 | 0.0561 (8) | 0.0355 (7) | 0.0608 (9) | −0.0017 (6) | −0.0169 (7) | −0.0092 (6) |
Geometric parameters (Å, º) top
| N3—C1 | 1.3593 (19) | N6—C5 | 1.314 (2) |
| N3—C2 | 1.3607 (18) | N2—C2 | 1.2966 (19) |
| N3—N5 | 1.3997 (18) | C5—N9 | 1.354 (2) |
| N8—C5 | 1.3576 (19) | C2—C3 | 1.492 (2) |
| N8—C4 | 1.362 (2) | N9—H9A | 0.88 (2) |
| N8—N10 | 1.4040 (19) | N9—H9B | 0.86 (3) |
| N1—C1 | 1.314 (2) | C4—C3 | 1.489 (2) |
| N1—N2 | 1.4094 (19) | N4—C1 | 1.341 (2) |
| N7—C4 | 1.297 (2) | N4—H4A | 0.87 (2) |
| N7—N6 | 1.4078 (19) | N4—H4B | 0.87 (2) |
| N10—H10B | 0.88 (3) | C3—H3A | 0.957 (18) |
| N10—H10A | 0.92 (3) | C3—H3B | 0.982 (19) |
| N5—H5B | 0.86 (3) | O1—H1A | 0.89 (3) |
| N5—H5A | 0.89 (3) | O1—H1B | 0.94 (3) |
| | | |
| C1—N3—C2 | 105.99 (12) | N3—C2—C3 | 122.99 (13) |
| C1—N3—N5 | 124.19 (12) | C5—N9—H9A | 114.2 (13) |
| C2—N3—N5 | 129.80 (13) | C5—N9—H9B | 117.9 (16) |
| C5—N8—C4 | 106.00 (12) | H9A—N9—H9B | 120 (2) |
| C5—N8—N10 | 124.31 (13) | N7—C4—N8 | 109.98 (13) |
| C4—N8—N10 | 129.53 (13) | N7—C4—C3 | 126.62 (14) |
| C1—N1—N2 | 106.37 (13) | N8—C4—C3 | 123.39 (13) |
| C4—N7—N6 | 107.58 (13) | C1—N4—H4A | 118.4 (13) |
| N8—N10—H10B | 109.4 (16) | C1—N4—H4B | 121.1 (15) |
| N8—N10—H10A | 106.1 (16) | H4A—N4—H4B | 121 (2) |
| H10B—N10—H10A | 106 (2) | N1—C1—N4 | 126.74 (15) |
| N3—N5—H5B | 109.2 (16) | N1—C1—N3 | 110.01 (13) |
| N3—N5—H5A | 109 (2) | N4—C1—N3 | 123.25 (14) |
| H5B—N5—H5A | 108 (3) | C4—C3—C2 | 111.50 (13) |
| C5—N6—N7 | 106.44 (12) | C4—C3—H3A | 108.8 (10) |
| C2—N2—N1 | 107.62 (12) | C2—C3—H3A | 109.3 (10) |
| N6—C5—N9 | 126.88 (15) | C4—C3—H3B | 109.5 (11) |
| N6—C5—N8 | 109.99 (13) | C2—C3—H3B | 108.9 (11) |
| N9—C5—N8 | 123.07 (15) | H3A—C3—H3B | 108.8 (14) |
| N2—C2—N3 | 110.01 (13) | H1A—O1—H1B | 106 (2) |
| N2—C2—C3 | 126.99 (13) | | |
| | | |
| C4—N7—N6—C5 | −0.31 (18) | N6—N7—C4—C3 | 178.91 (14) |
| C1—N1—N2—C2 | −0.66 (18) | C5—N8—C4—N7 | −0.05 (17) |
| N7—N6—C5—N9 | 177.54 (16) | N10—N8—C4—N7 | −175.58 (15) |
| N7—N6—C5—N8 | 0.29 (17) | C5—N8—C4—C3 | −178.79 (14) |
| C4—N8—C5—N6 | −0.16 (17) | N10—N8—C4—C3 | 5.7 (2) |
| N10—N8—C5—N6 | 175.67 (14) | N2—N1—C1—N4 | 179.14 (17) |
| C4—N8—C5—N9 | −177.53 (15) | N2—N1—C1—N3 | 0.13 (18) |
| N10—N8—C5—N9 | −1.7 (2) | C2—N3—C1—N1 | 0.41 (18) |
| N1—N2—C2—N3 | 0.93 (18) | N5—N3—C1—N1 | 178.92 (16) |
| N1—N2—C2—C3 | −178.11 (16) | C2—N3—C1—N4 | −178.64 (16) |
| C1—N3—C2—N2 | −0.85 (17) | N5—N3—C1—N4 | −0.1 (3) |
| N5—N3—C2—N2 | −179.24 (17) | N7—C4—C3—C2 | −113.06 (18) |
| C1—N3—C2—C3 | 178.24 (14) | N8—C4—C3—C2 | 65.5 (2) |
| N5—N3—C2—C3 | −0.1 (3) | N2—C2—C3—C4 | 9.6 (2) |
| N6—N7—C4—N8 | 0.22 (18) | N3—C2—C3—C4 | −169.34 (14) |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···N2 | 0.89 (3) | 2.04 (3) | 2.9308 (19) | 177 (2) |
| O1—H1B···N7i | 0.94 (3) | 1.92 (3) | 2.852 (2) | 172 (2) |
| N4—H4A···N1ii | 0.87 (2) | 2.09 (2) | 2.949 (2) | 170.1 (18) |
| N9—H9A···N6iii | 0.88 (2) | 2.16 (2) | 3.018 (2) | 163.6 (18) |
| N4—H4B···N5iv | 0.87 (2) | 2.35 (2) | 3.084 (2) | 142.4 (19) |
| N10—H10A···N6v | 0.92 (3) | 2.49 (3) | 3.369 (2) | 160 (2) |
| N10—H10B···O1v | 0.88 (3) | 2.33 (2) | 3.073 (2) | 142 (2) |
| Symmetry codes: (i) x−1, y, z; (ii) −x, −y+1, −z+1; (iii) −x+1, −y, −z; (iv) −x+1, −y+2, −z+1; (v) x, y+1, z. |
Nitrogen equivalents of different detonation products top| Detonation product | C | H2 | N2 | CO | CO2 |
| Nitrogen equivalent index | 0.15 | 0.29 | 1 | 0.78 | 1.35 |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
[email protected] for assistance.
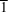 (triclinic) with Z = 2. The structure of BDATZM·H2O can be described as a two-dimensional ladder plane with extensive hydrogen bonding and no disorder. The thermal behaviour was studied under non-isothermal conditions by differential scanning calorimetry (DSC) and thermogravimetric/differential thermogravimetric (TG/DTG) methods. The detonation velocity (D) and detonation pressure (P) of BDATZM were estimated using the nitrogen equivalent equation according to the experimental density. A comparison between BDATZM·H2O and bis(5-amino-1,2,4-triazol-3-yl)methane (BATZM) was made to determine the effect of the amino group; the results suggest that the amino group increases the hydrophilicity, space utilization and energy, and decreases the thermal stability and symmetry of the resulting compound.
(triclinic) with Z = 2. The structure of BDATZM·H2O can be described as a two-dimensional ladder plane with extensive hydrogen bonding and no disorder. The thermal behaviour was studied under non-isothermal conditions by differential scanning calorimetry (DSC) and thermogravimetric/differential thermogravimetric (TG/DTG) methods. The detonation velocity (D) and detonation pressure (P) of BDATZM were estimated using the nitrogen equivalent equation according to the experimental density. A comparison between BDATZM·H2O and bis(5-amino-1,2,4-triazol-3-yl)methane (BATZM) was made to determine the effect of the amino group; the results suggest that the amino group increases the hydrophilicity, space utilization and energy, and decreases the thermal stability and symmetry of the resulting compound.
 journal menu
journal menu









 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry


