Buy article online - an online subscription or single-article purchase is required to access this article.
In the polymeric title compound, [CuI(C
10H
8N
4)]
n, the Cu
I atom is in a four-coordinated tetrahedral geometry, formed by two I atoms and two pyridine N atoms from two different 4,4'-(diazenediyl)dipyridine (4,4'-azpy) ligands. Two
![[mu]](/logos/entities/mu_rmgif.gif) 2
2-I atoms link two Cu
I atoms to form a planar rhomboid [Cu
2I
2] cluster located on an inversion centre, where the distance between two Cu
I atoms is 2.7781 (15) Å and the Cu-I bond lengths are 2.6290 (13) and 2.7495 (15) Å. The bridging 4,4'-azpy ligands connect the [Cu
2I
2] clusters into a two-dimensional (2-D) double-layered grid-like network [parallel to the (10
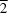
) plane], with a (4,4)-connected topology. Two 2-D grid-like networks interweave each other by long 4,4'-azpy bridging ligands to form a dense 2-D double-layered network. To the best of our knowledge, this interwoven 2-D
![[rightwards arrow]](/logos/entities/rarr_rmgif.gif)
2-D network is observed for the first time in [Cu
2I
2]-organic compounds.
Supporting information
CCDC reference: 925752
(NH4)2WOS3 (0.335 g, 1 mmol), CuSCN (0.568 g, 3 mmol) and
(t-Bu)4NI (1.119 g, 3 mmol) were added to dimethylformamide (DMF, 2 ml) and the resulting solution was stirred thoroughly for 5 min. After
filtration, i-PrOH (5 ml) was carefully layered on the surface of the
the red filtrate. Red block-shaped crystals were obtained after two weeks.
These crystals (0.0734 g) and 4,4'-(diazenediyl)dipyridine (0.0183 g, 0.1 mmol)
were added to acetonitrile (2 ml) and DMF (1 ml) and the resulting solution
was stirred thoroughly for 10 min in a vacuum. The solution was then sealed
and heated in an oven at 373 K for 48 h, and then cooled to room temperature
at a rate of 3 K h-1, producing brown block-shaped crystals (yield 0.0287 g). Analysis calculated for C10H8CuIN4: C 32.06, H 2.15, N 14.95%;
found: C 31.88, H 2.19, N 15.15%. IR (KBr, cm-1): 3053 (w), 2108
(s h), 1595 (s h), 1563 (w), 1413 (s h), 1224
(m), 1048 (w), 928 (s h), 841 (s h), 569
(m), 430 (m). [What does the "h" represent in the peak
assignments?]
H atoms were positioned geometrically and refined using a riding model, with
C—H = 0.93 Å and Uiso(H) = 1.2Ueq(C).
Data collection: CrystalClear (Rigaku, 2008); cell refinement: CrystalClear (Rigaku, 2008); data reduction: CrystalClear (Rigaku, 2008); program(s) used to solve structure: SHELXTL (Sheldrick, 2008); program(s) used to refine structure: SHELXTL (Sheldrick, 2008); molecular graphics: SHELXTL (Sheldrick, 2008); software used to prepare material for publication: SHELXTL (Sheldrick, 2008).
poly[[µ-4,4'-(diazenediyl)dipyridine]-µ
2-iodido-copper(I)]
top
Crystal data top
| [CuI(C10H8N4)] | F(000) = 712 |
| Mr = 374.65 | Dx = 2.085 Mg m−3 |
| Monoclinic, P2/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yc | Cell parameters from 3410 reflections |
| a = 9.7137 (19) Å | θ = 2.5–28.9° |
| b = 8.0745 (16) Å | µ = 4.39 mm−1 |
| c = 15.396 (3) Å | T = 150 K |
| β = 98.73 (3)° | Block, brown |
| V = 1193.6 (4) Å3 | 0.2 × 0.18 × 0.15 mm |
| Z = 4 | |
Data collection top
Rigaku Saturn724+ (2x2 bin mode)
diffractometer | 2171 independent reflections |
| Radiation source: fine-focus sealed tube | 1674 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.053 |
| ω scans | θmax = 25.3°, θmin = 2.9° |
Absorption correction: multi-scan
(CrystalClear; Rigaku, 2008) | h = −11→9 |
| Tmin = 0.795, Tmax = 1.000 | k = −9→8 |
| 6682 measured reflections | l = −17→18 |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.060 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.093 | H-atom parameters constrained |
| S = 1.17 | w = 1/[σ2(Fo2) + (0.0198P)2 + 2.6592P]
where P = (Fo2 + 2Fc2)/3 |
| 2171 reflections | (Δ/σ)max < 0.001 |
| 145 parameters | Δρmax = 0.80 e Å−3 |
| 0 restraints | Δρmin = −0.63 e Å−3 |
Crystal data top
| [CuI(C10H8N4)] | V = 1193.6 (4) Å3 |
| Mr = 374.65 | Z = 4 |
| Monoclinic, P2/c | Mo Kα radiation |
| a = 9.7137 (19) Å | µ = 4.39 mm−1 |
| b = 8.0745 (16) Å | T = 150 K |
| c = 15.396 (3) Å | 0.2 × 0.18 × 0.15 mm |
| β = 98.73 (3)° | |
Data collection top
Rigaku Saturn724+ (2x2 bin mode)
diffractometer | 2171 independent reflections |
Absorption correction: multi-scan
(CrystalClear; Rigaku, 2008) | 1674 reflections with I > 2σ(I) |
| Tmin = 0.795, Tmax = 1.000 | Rint = 0.053 |
| 6682 measured reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.060 | 0 restraints |
| wR(F2) = 0.093 | H-atom parameters constrained |
| S = 1.17 | Δρmax = 0.80 e Å−3 |
| 2171 reflections | Δρmin = −0.63 e Å−3 |
| 145 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | |
| I1 | 0.10063 (6) | 0.31252 (6) | 0.41593 (4) | 0.0507 (2) | |
| Cu1 | 0.05260 (10) | 0.62373 (12) | 0.45162 (7) | 0.0496 (3) | |
| N1 | −0.0745 (6) | 0.7213 (8) | 0.3467 (4) | 0.0404 (16) | |
| N2 | 0.2398 (6) | 0.7250 (8) | 0.5042 (4) | 0.0440 (17) | |
| N3 | 0.5868 (7) | 0.9279 (10) | 0.6661 (4) | 0.056 (2) | |
| N4 | 0.6006 (7) | 1.0804 (10) | 0.6633 (4) | 0.0525 (19) | |
| C1 | −0.0876 (8) | 0.8843 (10) | 0.3362 (5) | 0.046 (2) | |
| H1A | −0.0247 | 0.9520 | 0.3713 | 0.056* | |
| C2 | −0.1888 (8) | 0.9597 (10) | 0.2763 (5) | 0.050 (2) | |
| H2A | −0.1931 | 1.0744 | 0.2708 | 0.059* | |
| C3 | −0.2833 (8) | 0.8588 (10) | 0.2249 (5) | 0.042 (2) | |
| C4 | −0.2708 (9) | 0.6913 (11) | 0.2340 (6) | 0.068 (3) | |
| H4A | −0.3336 | 0.6213 | 0.2003 | 0.081* | |
| C5 | −0.1640 (8) | 0.6267 (11) | 0.2936 (5) | 0.060 (3) | |
| H5A | −0.1536 | 0.5123 | 0.2971 | 0.072* | |
| C6 | 0.2540 (8) | 0.8884 (10) | 0.5123 (5) | 0.048 (2) | |
| H6A | 0.1845 | 0.9550 | 0.4823 | 0.057* | |
| C7 | 0.3652 (8) | 0.9648 (10) | 0.5625 (5) | 0.050 (2) | |
| H7A | 0.3699 | 1.0796 | 0.5665 | 0.060* | |
| C8 | 0.4689 (7) | 0.8682 (11) | 0.6067 (5) | 0.045 (2) | |
| C9 | 0.4586 (9) | 0.6984 (11) | 0.5976 (6) | 0.073 (3) | |
| H9A | 0.5280 | 0.6294 | 0.6259 | 0.087* | |
| C10 | 0.3424 (8) | 0.6326 (11) | 0.5452 (6) | 0.069 (3) | |
| H10A | 0.3363 | 0.5182 | 0.5386 | 0.083* | |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| I1 | 0.0569 (4) | 0.0338 (3) | 0.0588 (4) | 0.0023 (3) | 0.0007 (3) | −0.0019 (3) |
| Cu1 | 0.0447 (6) | 0.0386 (6) | 0.0579 (7) | −0.0023 (5) | −0.0164 (5) | 0.0030 (5) |
| N1 | 0.041 (4) | 0.031 (4) | 0.047 (4) | 0.000 (3) | −0.001 (3) | −0.001 (3) |
| N2 | 0.039 (4) | 0.035 (4) | 0.052 (4) | 0.000 (3) | −0.010 (3) | 0.001 (3) |
| N3 | 0.041 (4) | 0.062 (5) | 0.063 (5) | −0.010 (4) | −0.005 (4) | −0.008 (4) |
| N4 | 0.043 (4) | 0.061 (5) | 0.050 (4) | −0.010 (4) | −0.005 (3) | −0.005 (4) |
| C1 | 0.045 (5) | 0.044 (6) | 0.046 (5) | −0.003 (4) | −0.008 (4) | −0.004 (4) |
| C2 | 0.054 (5) | 0.040 (5) | 0.052 (5) | 0.011 (4) | 0.000 (4) | 0.009 (4) |
| C3 | 0.044 (5) | 0.042 (5) | 0.036 (4) | 0.009 (4) | −0.004 (4) | 0.002 (4) |
| C4 | 0.058 (6) | 0.057 (6) | 0.074 (6) | 0.004 (5) | −0.035 (5) | −0.003 (5) |
| C5 | 0.061 (6) | 0.037 (5) | 0.072 (6) | 0.003 (4) | −0.020 (5) | −0.001 (4) |
| C6 | 0.048 (5) | 0.041 (6) | 0.049 (5) | −0.003 (4) | −0.008 (4) | 0.002 (4) |
| C7 | 0.051 (5) | 0.047 (6) | 0.048 (5) | −0.016 (4) | −0.005 (4) | −0.003 (4) |
| C8 | 0.030 (4) | 0.060 (6) | 0.044 (5) | 0.000 (4) | −0.001 (4) | −0.002 (4) |
| C9 | 0.058 (6) | 0.043 (6) | 0.102 (8) | 0.009 (5) | −0.035 (6) | −0.018 (5) |
| C10 | 0.053 (6) | 0.041 (6) | 0.101 (7) | 0.000 (4) | −0.032 (5) | −0.016 (5) |
Geometric parameters (Å, º) top
| I1—Cu1 | 2.6290 (13) | C2—H2A | 0.9300 |
| I1—Cu1i | 2.7495 (15) | C3—C4 | 1.364 (11) |
| Cu1—N1 | 2.037 (6) | C3—N4iii | 1.444 (9) |
| Cu1—N2 | 2.045 (6) | C4—C5 | 1.379 (10) |
| N1—C1 | 1.330 (9) | C4—H4A | 0.9300 |
| N1—C5 | 1.338 (9) | C5—H5A | 0.9300 |
| N2—C10 | 1.326 (9) | C6—C7 | 1.375 (9) |
| N2—C6 | 1.330 (9) | C6—H6A | 0.9300 |
| N3—N4 | 1.240 (9) | C7—C8 | 1.370 (10) |
| N3—C8 | 1.435 (9) | C7—H7A | 0.9300 |
| N4—C3ii | 1.444 (9) | C8—C9 | 1.381 (11) |
| C1—C2 | 1.382 (9) | C9—C10 | 1.389 (10) |
| C1—H1A | 0.9300 | C9—H9A | 0.9300 |
| C2—C3 | 1.383 (10) | C10—H10A | 0.9300 |
| | | |
| Cu1—I1—Cu1i | 62.15 (4) | C4—C3—C2 | 119.0 (7) |
| N1—Cu1—N2 | 123.7 (3) | C4—C3—N4iii | 117.0 (7) |
| N1—Cu1—I1 | 108.02 (18) | C2—C3—N4iii | 124.0 (7) |
| N2—Cu1—I1 | 106.82 (19) | C3—C4—C5 | 119.3 (7) |
| N1—Cu1—I1i | 100.64 (19) | C3—C4—H4A | 120.3 |
| N2—Cu1—I1i | 100.42 (19) | C5—C4—H4A | 120.3 |
| I1—Cu1—I1i | 117.85 (4) | N1—C5—C4 | 122.9 (8) |
| N1—Cu1—Cu1i | 118.39 (18) | N1—C5—H5A | 118.5 |
| N2—Cu1—Cu1i | 116.93 (18) | C4—C5—H5A | 118.5 |
| I1—Cu1—Cu1i | 61.05 (4) | N2—C6—C7 | 124.0 (7) |
| I1i—Cu1—Cu1i | 56.80 (4) | N2—C6—H6A | 118.0 |
| C1—N1—C5 | 116.7 (6) | C7—C6—H6A | 118.0 |
| C1—N1—Cu1 | 121.0 (5) | C8—C7—C6 | 118.6 (8) |
| C5—N1—Cu1 | 121.5 (5) | C8—C7—H7A | 120.7 |
| C10—N2—C6 | 116.9 (6) | C6—C7—H7A | 120.7 |
| C10—N2—Cu1 | 121.5 (5) | C7—C8—C9 | 118.6 (7) |
| C6—N2—Cu1 | 120.5 (5) | C7—C8—N3 | 125.5 (8) |
| N4—N3—C8 | 113.1 (7) | C9—C8—N3 | 115.9 (7) |
| N3—N4—C3ii | 113.1 (7) | C8—C9—C10 | 118.6 (7) |
| N1—C1—C2 | 124.2 (7) | C8—C9—H9A | 120.7 |
| N1—C1—H1A | 117.9 | C10—C9—H9A | 120.7 |
| C2—C1—H1A | 117.9 | N2—C10—C9 | 123.2 (8) |
| C1—C2—C3 | 117.7 (7) | N2—C10—H10A | 118.4 |
| C1—C2—H2A | 121.1 | C9—C10—H10A | 118.4 |
| C3—C2—H2A | 121.1 | | |
| | | |
| Cu1i—I1—Cu1—N1 | −113.0 (2) | Cu1—N1—C1—C2 | −168.1 (6) |
| Cu1i—I1—Cu1—N2 | 111.9 (2) | N1—C1—C2—C3 | 0.8 (13) |
| Cu1i—I1—Cu1—I1i | 0.0 | C1—C2—C3—C4 | −1.5 (13) |
| N2—Cu1—N1—C1 | −37.0 (7) | C1—C2—C3—N4iii | 176.3 (7) |
| I1—Cu1—N1—C1 | −162.6 (6) | C2—C3—C4—C5 | −0.4 (14) |
| I1i—Cu1—N1—C1 | 73.3 (6) | N4iii—C3—C4—C5 | −178.3 (8) |
| Cu1i—Cu1—N1—C1 | 131.1 (6) | C1—N1—C5—C4 | −3.8 (13) |
| N2—Cu1—N1—C5 | 153.7 (6) | Cu1—N1—C5—C4 | 166.0 (7) |
| I1—Cu1—N1—C5 | 28.0 (7) | C3—C4—C5—N1 | 3.2 (15) |
| I1i—Cu1—N1—C5 | −96.1 (6) | C10—N2—C6—C7 | −2.2 (13) |
| Cu1i—Cu1—N1—C5 | −38.2 (7) | Cu1—N2—C6—C7 | 166.0 (6) |
| N1—Cu1—N2—C10 | −152.1 (7) | N2—C6—C7—C8 | 0.5 (13) |
| I1—Cu1—N2—C10 | −26.0 (7) | C6—C7—C8—C9 | 1.2 (13) |
| I1i—Cu1—N2—C10 | 97.5 (7) | C6—C7—C8—N3 | −175.7 (8) |
| Cu1i—Cu1—N2—C10 | 39.6 (8) | N4—N3—C8—C7 | −11.7 (12) |
| N1—Cu1—N2—C6 | 40.2 (7) | N4—N3—C8—C9 | 171.4 (9) |
| I1—Cu1—N2—C6 | 166.3 (6) | C7—C8—C9—C10 | −1.0 (14) |
| I1i—Cu1—N2—C6 | −70.2 (6) | N3—C8—C9—C10 | 176.2 (9) |
| Cu1i—Cu1—N2—C6 | −128.1 (6) | C6—N2—C10—C9 | 2.4 (14) |
| C8—N3—N4—C3ii | 178.2 (6) | Cu1—N2—C10—C9 | −165.7 (8) |
| C5—N1—C1—C2 | 1.8 (12) | C8—C9—C10—N2 | −0.8 (16) |
| Symmetry codes: (i) −x, −y+1, −z+1; (ii) x+1, −y+2, z+1/2; (iii) x−1, −y+2, z−1/2. |
Experimental details
| Crystal data |
| Chemical formula | [CuI(C10H8N4)] |
| Mr | 374.65 |
| Crystal system, space group | Monoclinic, P2/c |
| Temperature (K) | 150 |
| a, b, c (Å) | 9.7137 (19), 8.0745 (16), 15.396 (3) |
| β (°) | 98.73 (3) |
| V (Å3) | 1193.6 (4) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 4.39 |
| Crystal size (mm) | 0.2 × 0.18 × 0.15 |
| |
| Data collection |
| Diffractometer | Rigaku Saturn724+ (2x2 bin mode)
diffractometer |
| Absorption correction | Multi-scan
(CrystalClear; Rigaku, 2008) |
| Tmin, Tmax | 0.795, 1.000 |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 6682, 2171, 1674 |
| Rint | 0.053 |
| (sin θ/λ)max (Å−1) | 0.602 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.060, 0.093, 1.17 |
| No. of reflections | 2171 |
| No. of parameters | 145 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.80, −0.63 |

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry
The full text of this article is available to subscribers to the journal.
If you have already registered and are using a computer listed in your registration details, please email
[email protected] for assistance.
![[mu]](/logos/entities/mu_rmgif.gif) 2-I atoms link two CuI atoms to form a planar rhomboid [Cu2I2] cluster located on an inversion centre, where the distance between two CuI atoms is 2.7781 (15) Å and the Cu-I bond lengths are 2.6290 (13) and 2.7495 (15) Å. The bridging 4,4'-azpy ligands connect the [Cu2I2] clusters into a two-dimensional (2-D) double-layered grid-like network [parallel to the (10
2-I atoms link two CuI atoms to form a planar rhomboid [Cu2I2] cluster located on an inversion centre, where the distance between two CuI atoms is 2.7781 (15) Å and the Cu-I bond lengths are 2.6290 (13) and 2.7495 (15) Å. The bridging 4,4'-azpy ligands connect the [Cu2I2] clusters into a two-dimensional (2-D) double-layered grid-like network [parallel to the (10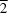 ) plane], with a (4,4)-connected topology. Two 2-D grid-like networks interweave each other by long 4,4'-azpy bridging ligands to form a dense 2-D double-layered network. To the best of our knowledge, this interwoven 2-D
) plane], with a (4,4)-connected topology. Two 2-D grid-like networks interweave each other by long 4,4'-azpy bridging ligands to form a dense 2-D double-layered network. To the best of our knowledge, this interwoven 2-D![[rightwards arrow]](/logos/entities/rarr_rmgif.gif) 2-D network is observed for the first time in [Cu2I2]-organic compounds.
2-D network is observed for the first time in [Cu2I2]-organic compounds.![[mu]](/logos/entities/mu_rmgif.gif) 2-I atoms link two CuI atoms to form a planar rhomboid [Cu2I2] cluster located on an inversion centre, where the distance between two CuI atoms is 2.7781 (15) Å and the Cu-I bond lengths are 2.6290 (13) and 2.7495 (15) Å. The bridging 4,4'-azpy ligands connect the [Cu2I2] clusters into a two-dimensional (2-D) double-layered grid-like network [parallel to the (10
2-I atoms link two CuI atoms to form a planar rhomboid [Cu2I2] cluster located on an inversion centre, where the distance between two CuI atoms is 2.7781 (15) Å and the Cu-I bond lengths are 2.6290 (13) and 2.7495 (15) Å. The bridging 4,4'-azpy ligands connect the [Cu2I2] clusters into a two-dimensional (2-D) double-layered grid-like network [parallel to the (10![[rightwards arrow]](/logos/entities/rarr_rmgif.gif) 2-D network is observed for the first time in [Cu2I2]-organic compounds.
2-D network is observed for the first time in [Cu2I2]-organic compounds.
 journal menu
journal menu








![[Figure 1]](https://journals.iucr.org/c/issues/2013/02/00/sk3470/sk3470fig1thm.gif)
![[Figure 2]](https://journals.iucr.org/c/issues/2013/02/00/sk3470/sk3470fig2thm.gif)
![[Figure 3]](https://journals.iucr.org/c/issues/2013/02/00/sk3470/sk3470fig3thm.gif)

 Subscribe to Acta Crystallographica Section C: Structural Chemistry
Subscribe to Acta Crystallographica Section C: Structural Chemistry



In recent years, the construction of multidimensional cluster-based coordination polymers have been a major focus of research in the field of chemistry and materials science, due to not only their aesthetically intriguing structural features (Schubert, 2011; Zhang, Song et al., 2008) but also because of their potentially useful applications, such as magnetism (Imai et al., 2009), photoluminescence (Perruchas et al., 2011), ion exchange (Mrutu et al., 2011), catalysis (Tan et al., 2012), adsorption (Li et al., 2010) and nonlinear optical properties (Zhang, Cao et al., 2008; Zhang, Meng et al., 2010). The strategy popularly used is a building-block approach (Hu et al., 2005; Eddaoudi et al., 2001) and much effort has been devoted to the connection of suitable predetermined building blocks into frameworks in order to obtain desired structures and properties.
As an important kind of cluster, copper(I) halide clusters have been widely studied due to their diverse structures and applications (Peng et al., 2010; Zhang, Wu et al., 2010). Up to now, copper(I) halide cluster-based coordination polymers with various structural motifs have been constructed, such as rhomboid [Cu2I2] dimers (Niu et al., 2006; Araki et al., 2005), cubane-like [Cu4I4] tetramers (Blake et al., 2001; Chen et al., 2008), zigzag chains (Graham et al., 2000; Cheng et al., 2004), double-stranded ladders (Graham et al., 2000) and two-dimensional [CuI]n layers (Peng et al., 2005). Various bridging ligands, such as phosphines, anilines, pyridine-type ligands and thioethers, have been applied in constructing CuX-based coordination polymers (Peng et al., 2010). Herein, we have used 4,4'-(diazenediyl)dipyridine (4,4'-azpy) ligands as long bridges to construct the CuI-based coordination polymer poly[[µ-4,4'-(diazenediyl)dipyridine]-µ2-iodido-copper(I)], (I), which is formed by [Cu2I2] clusters and single 4,4'-azpy bridges, and exhibits a novel twofold interwoven two-dimensional → two-dimensional (2-D → 2-D) network.
X-ray crystallographic analysis revealed that (I) crystallizes in the monoclinic space group P2/c with each [Cu2I2] cluster linked to four other [Cu2I2] clusters through long single 4,4'-azpy bridges, furnishing a 2-D double-layered grid-like network with a (4,4)-connected topology (Figs. 2 and 3). As shown in Fig. 1, the CuI atom is bonded by two I atoms and two pyridine N atoms from two 4,4'-azpy ligands, forming a four-coordinated tetrahedral geometry. The Cu1—N1 and Cu1—N2 bond lengths are 2.037 (6) and 2.045 (6) Å, respectively, and the N1—Cu1—N2 angle is 123.7 (3)°. The planar rhomboid [Cu2I2] cluster is formed by two µ2-I atoms linking two CuI atoms. In the [Cu2I2] cluster, the distance between two CuI atoms is 2.7781 (15) Å, and the Cu—I bonds lengths are 2.6290 (13) and 2.7495 (15) Å. The I—Cu—I and Cu—I—Cu angles are 117.85 (4) and 62.15 (4)°, respectively.
The 4,4'-azpy ligands connect the [Cu2I2] clusters into a 2-D double-layered grid-like network with a (4,4)-connected topology. The N═N bond length is 1.240 (9) Å and the average distances between C and N atoms in the pyridine rings is 1.331 (9) Å. In the (4,4) network, the [Cu2I2] clusters shows a unique ABAB arrangement (Fig. 2) and the 4,4'-azpy ligand is slightly distorted because the angle between the planes of the two pyridine is 8.9 (4)°. Interestingly, the 2-D net exhibits a rare double-layered structure (Fig. 3a), which should be due to the distances between the two CuI atoms in the [Cu2I2] clusters. The lengths of the two diagonals in the grid, constructed by four [Cu2I2] clusters and four 4,4'-azpy bridges, are 16.149 (2) and 22.883 (3) Å (Fig. 2). Therefore, each layer has enough empty space, which results in two layers interweaving each other by long 4,4'-azpy bridging ligands to form a new dense two-dimensional double-layered network (Fig. 3b).
In previous work, the phenomena of interpenetration and catenation have appeared in CuX-based coordination polymers. In [Cu2Br2{1,2-bis(pyridin-4-yl)ethane}2]n (Hu et al., 2006), two sets of (4,4) layers adopt the diagonal–diagonal fourfold interpenetration mode, giving high catenation and generating a 2-D → 3-D (three-dimensional) interpenetrating framework. In [Cu2Br2{1,3-bis(pyridin-4-yl)propane}2]n (Hu et al., 2006), the 2-D polycatenane network is formed by interlocking of 1-D (one-dimensional) double-stranded tubular chains. However, interwoven 2-D → 2-D networks are very scarce in coordination polymers and, to the best of our knowledge, the novel interwoven 2-D → 2-D network in (I) is the first to be found in a [Cu2X2]–organic compound.
In fact, this experiment was designed to synthesize W/S/Cu cluster-based coordination polymers. In the first step of the preparation, the cubane-shaped {[(t-Bu)4N]3[WOS3(CuI)3I]} W/S/Cu cluster was synthesized. It was hoped that in the next step the W/S/Cu clusters would be bridged by long 4,4'-azpy ligands to form a cubane-shaped W/S/Cu cluster-based coordination polymer. However, in the second step, the W/S/Cu clusters were decomposed by water from the acetonitrile or dimethylformamide solvent in the solvothermal reaction. As a result, the unexpected [Cu2I2]-based coordination polymer (I) was obtained.
In summary, an unexpected [Cu2I2]-based coordination polymer, (I), with a two-dimensional double-layered grid-like (4,4) network is obtained from a solvothermal reaction which was originally designed to synthesize W/S/Cu cluster-based coordination polymers. This interwoven two-dimensional → two-dimensional network in (I) is firstly found in [Cu2I2]-organic compounds.