
The title porphyrin compound forms hydrogen-bonded adducts with methanol (1:1), [Pd(C
48H
28N
4O
8)]·CH
4O, (I), and with water and
N,
N-dimethylformamide (1:4:4), [Pd(C
48H
28N
4O
8)]·4C
3H
7NO·4H
2O, (II). In (I), the metalloporphyrin unit lies across a mirror plane in
Cmca, while in (II), this unit lies across an inversion center in
P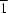
. Extended supramolecular hydrogen-bonded arrays are formed in (I) by intermolecular interactions between the carboxylic acid functions, either directly or through the methanol species. These layers have a wavy topology and large interporphyrin pores, which are filled in the crystal structure by double interpenetration as well as enclathration of additional non-interacting nitrobenzene solvent molecules. The supramolecular aggregation in (II) can be characterized by cascaded porphyrin layers, wherein adjacent porphyrin molecules are hydrogen bonded to one another through molecules of water that are incorporated into the hydrogen-bonding scheme. Molecules of dimethylformamide partly solvate the carboxylic acid groups and fill the interporphyrin space in the crystal structure.
Supporting information
CCDC references: 677162; 677163
Pd-TCPP was obtained commercially from Porphyrin Systems GbR. Compound (I) was
obtained by dissolving the metalloporphyrin (2.7 mg, 0.003 mmol) in a minimal
amount of methanol. To this, six drops of nitrobenzene were added. Slow
evaporation of the resulting mixture yielded after one month X-ray quality red
rhombus-like crystals. Reaction of Pd-TCPP (2 mg, 0.002 mmol) with a minimal
amount of methanol and a few drops of DMF yielded crystals of (II) under
otherwise similar crystallization conditions.
H atoms bound to C atoms were located in calculated positions and were
constrained to ride on their parent atoms with C—H distances of 0.95 and
0.98 Å and with Uiso(H) values of 1.2 or 1.5 times Ueq(C).
H atoms bound to O atoms were either located in difference Fourier maps or
placed in calculated positions to correspond to idealized hydrogen-bonding
geometries, with O—H distances within the range 0.84–0.99 Å. Their atomic
positions were not refined, and they were constrained to ride on their parent
atoms with Uiso(H) values of 1.2 or 1.5 times Ueq(O)
[should this be just 1.2?]. In (I), the porphyrin and methanol
components are located on mirror planes and twofold axes, respectively. The
hydrogen-bonding scheme is characterized by a twofold disorder about the
rotation axes. Correspondingly, the methanol molecule and the carboxylic acid
groups also exhibit partial disorder. The nitrobenzene solvent incorporated
into the crystal structure of (I) is also positioned on, and severely
disordered about, the (1/2 y z) mirror plane, being centered
approximately at (1/2, 0.1, 0.4). It could not be reliably modeled by discrete
atoms. Correspondingly, its contribution was subtracted from the diffraction
data by the SQUEEZE procedure (PLATON; Spek, 2003). In (II), one of the
DMF species (O39/C40/N41/C42/C43) exhibits a twofold orientational disorder,
which could be modeled, and it was refined with restrained geometry in order
to avoid irregular values for bond lengths and bond angles. The Pd-TCPP
reference unit is located on an inversion centre at (0, 0, 1).
For both compounds, data collection: Collect (Nonius, 1999); cell refinement: DENZO (Otwinowski & Minor, 1997). Data reduction: DENZO and SCALEPACK (Otwinowski & Minor, 1997) for (I); DENZO (Otwinowski & Minor, 1997) for (II). For both compounds, program(s) used to solve structure: SIR97 (Altomare et al., 1999); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: ORTEPIII (Burnett & Johnson, 1996) and Mercury (Macrae et al., 2006); software used to prepare material for publication: SHELXL97 (Sheldrick, 1997).
(I) [5,10,15,20-tetrakis(4-carboxyphenyl)porphyrinato]palladium(II) methanol solvate
top
Crystal data top
| [Pd(C48H28N4O8)]·CH4O | F(000) = 3776 |
| Mr = 927.21 | Dx = 1.354 Mg m−3 |
| Orthorhombic, Cmca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -C 2bc 2 | Cell parameters from 4588 reflections |
| a = 31.0347 (6) Å | θ = 2.6–25.6° |
| b = 15.9441 (11) Å | µ = 0.47 mm−1 |
| c = 18.3824 (7) Å | T = 110 K |
| V = 9096.0 (7) Å3 | Rhomb, red |
| Z = 8 | 0.30 × 0.10 × 0.05 mm |
Data collection top
Nonius KappaCCD
diffractometer | 2688 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.098 |
| Graphite monochromator | θmax = 25.6°, θmin = 2.6° |
| Detector resolution: 12.8 pixels mm-1 | h = 0→37 |
| 0.5 deg. ϕ scans | k = 0→19 |
| 20365 measured reflections | l = −22→0 |
| 4344 independent reflections | |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.070 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.160 | H-atom parameters constrained |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0724P)2 + 2.4049P]
where P = (Fo2 + 2Fc2)/3 |
| 4344 reflections | (Δ/σ)max = 0.008 |
| 291 parameters | Δρmax = 0.50 e Å−3 |
| 12 restraints | Δρmin = −0.77 e Å−3 |
Crystal data top
| [Pd(C48H28N4O8)]·CH4O | V = 9096.0 (7) Å3 |
| Mr = 927.21 | Z = 8 |
| Orthorhombic, Cmca | Mo Kα radiation |
| a = 31.0347 (6) Å | µ = 0.47 mm−1 |
| b = 15.9441 (11) Å | T = 110 K |
| c = 18.3824 (7) Å | 0.30 × 0.10 × 0.05 mm |
Data collection top
Nonius KappaCCD
diffractometer | 2688 reflections with I > 2σ(I) |
| 20365 measured reflections | Rint = 0.098 |
| 4344 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.070 | 12 restraints |
| wR(F2) = 0.160 | H-atom parameters constrained |
| S = 1.00 | Δρmax = 0.50 e Å−3 |
| 4344 reflections | Δρmin = −0.77 e Å−3 |
| 291 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. The crystal contains severely disordered molecules of the nitrobenzene solvent,
which couldn't be modeled reliably by discrete atoms. One molecule of
nitrobenzene is located on and disordered about the mirror plane at
1/2,y,z, being centered at 1/2,0.1,0.4. Correspondingly, the
contribution of the disordered solvent was subtracted from the diffraction
data by the SQUEEZE procedure (Spek, 2003). The results below are based on
thus modified intensity data. The two independent carboxylic groups also
reveal partial rotational disorder about the C-COOH bond and SIMU restratrains
were applied to their ADP's. The methanol is also partly disordered. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Pd1 | 0.0000 | 0.03589 (4) | 0.35956 (3) | 0.0237 (2) | |
| C2 | 0.02189 (18) | 0.1673 (4) | 0.5582 (3) | 0.0309 (14) | |
| H2 | 0.0401 | 0.1876 | 0.5958 | 0.037* | |
| C3 | 0.03551 (18) | 0.1305 (4) | 0.4903 (3) | 0.0293 (14) | |
| C4 | 0.07828 (17) | 0.1262 (4) | 0.4667 (3) | 0.0255 (13) | |
| C5 | 0.09124 (18) | 0.0853 (4) | 0.4028 (3) | 0.0265 (13) | |
| C6 | 0.13500 (17) | 0.0824 (4) | 0.3773 (3) | 0.0309 (15) | |
| H6 | 0.1591 | 0.1099 | 0.3983 | 0.037* | |
| C7 | 0.13524 (17) | 0.0327 (4) | 0.3172 (3) | 0.0311 (14) | |
| H7 | 0.1598 | 0.0178 | 0.2891 | 0.037* | |
| C8 | 0.09142 (18) | 0.0067 (4) | 0.3040 (3) | 0.0283 (14) | |
| C9 | 0.07875 (17) | −0.0460 (4) | 0.2478 (3) | 0.0278 (13) | |
| C10 | 0.03559 (18) | −0.0646 (4) | 0.2321 (3) | 0.0294 (14) | |
| C11 | 0.02210 (18) | −0.1147 (4) | 0.1706 (3) | 0.0334 (15) | |
| H11 | 0.0403 | −0.1423 | 0.1367 | 0.040* | |
| N12 | 0.0000 | 0.1062 (5) | 0.4506 (3) | 0.0271 (16) | |
| N13 | 0.06496 (13) | 0.0403 (3) | 0.3576 (2) | 0.0258 (10) | |
| N14 | 0.0000 | −0.0358 (5) | 0.2700 (3) | 0.0264 (15) | |
| C15 | 0.11126 (17) | 0.1686 (4) | 0.5126 (3) | 0.0281 (14) | |
| C16 | 0.11155 (18) | 0.2557 (4) | 0.5201 (3) | 0.0327 (15) | |
| H16 | 0.0915 | 0.2887 | 0.4935 | 0.039* | |
| C17 | 0.14104 (19) | 0.2949 (5) | 0.5662 (3) | 0.0405 (17) | |
| H17 | 0.1420 | 0.3543 | 0.5695 | 0.049* | |
| C18 | 0.16916 (18) | 0.2456 (5) | 0.6075 (3) | 0.0360 (15) | |
| C19 | 0.1693 (2) | 0.1591 (5) | 0.6009 (3) | 0.0436 (18) | |
| H19 | 0.1889 | 0.1262 | 0.6285 | 0.052* | |
| C20 | 0.14048 (19) | 0.1207 (5) | 0.5534 (3) | 0.0380 (16) | |
| H20 | 0.1406 | 0.0614 | 0.5486 | 0.046* | |
| C21 | 0.1996 (2) | 0.2884 (5) | 0.6601 (3) | 0.0395 (17) | |
| O22 | 0.19883 (16) | 0.3654 (4) | 0.6649 (4) | 0.091 (2) | |
| H22 | 0.2156 | 0.3929 | 0.6926 | 0.109* | 0.50 |
| O23 | 0.22375 (15) | 0.2398 (4) | 0.6969 (2) | 0.0738 (19) | |
| H23 | 0.2422 | 0.2624 | 0.7237 | 0.089* | 0.50 |
| C24 | 0.11290 (17) | −0.0826 (4) | 0.2009 (3) | 0.0300 (14) | |
| C25 | 0.1340 (2) | −0.0354 (5) | 0.1482 (3) | 0.0462 (16) | |
| H25 | 0.1268 | 0.0220 | 0.1416 | 0.055* | |
| C26 | 0.1659 (2) | −0.0728 (5) | 0.1048 (4) | 0.0461 (19) | |
| H26 | 0.1803 | −0.0400 | 0.0691 | 0.055* | |
| C27 | 0.17659 (19) | −0.1556 (5) | 0.1130 (4) | 0.0385 (16) | |
| C28 | 0.1563 (2) | −0.2028 (5) | 0.1647 (4) | 0.0445 (17) | |
| H28 | 0.1637 | −0.2602 | 0.1706 | 0.053* | |
| C29 | 0.12447 (19) | −0.1666 (5) | 0.2095 (3) | 0.0388 (17) | |
| H29 | 0.1108 | −0.1996 | 0.2458 | 0.047* | |
| C30 | 0.2105 (2) | −0.1966 (5) | 0.0644 (3) | 0.0391 (16) | |
| O31 | 0.23083 (13) | −0.1452 (3) | 0.0237 (2) | 0.0509 (13) | |
| H31 | 0.2487 | −0.1723 | −0.0063 | 0.061* | |
| O32 | 0.21594 (14) | −0.2735 (3) | 0.0659 (2) | 0.0475 (13) | |
| C33 | 0.2500 | −0.0366 (8) | 0.2500 | 0.077 (4) | |
| H33A | 0.2797 | −0.0571 | 0.2530 | 0.116* | 0.50 |
| H33B | 0.2367 | −0.0571 | 0.2051 | 0.116* | 0.50 |
| H33C | 0.2336 | −0.0571 | 0.2920 | 0.116* | 0.50 |
| O34 | 0.2500 | 0.0498 (6) | 0.2500 | 0.124 (4) | |
| H34 | 0.2677 | 0.0787 | 0.2795 | 0.149* | 0.50 |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Pd1 | 0.0213 (3) | 0.0260 (4) | 0.0238 (3) | 0.000 | 0.000 | −0.0054 (3) |
| C2 | 0.033 (3) | 0.030 (4) | 0.030 (3) | −0.008 (3) | −0.007 (2) | −0.003 (3) |
| C3 | 0.034 (3) | 0.035 (4) | 0.019 (3) | −0.006 (3) | −0.006 (3) | −0.003 (3) |
| C4 | 0.027 (3) | 0.028 (4) | 0.021 (3) | −0.001 (3) | −0.003 (2) | −0.006 (2) |
| C5 | 0.028 (3) | 0.024 (4) | 0.027 (3) | 0.001 (3) | 0.000 (2) | 0.000 (3) |
| C6 | 0.021 (3) | 0.031 (4) | 0.040 (4) | −0.003 (3) | −0.002 (3) | 0.000 (3) |
| C7 | 0.023 (3) | 0.037 (4) | 0.033 (3) | −0.001 (3) | 0.012 (2) | −0.004 (3) |
| C8 | 0.028 (3) | 0.027 (4) | 0.030 (3) | 0.002 (3) | −0.006 (3) | 0.002 (3) |
| C9 | 0.033 (3) | 0.024 (4) | 0.027 (3) | −0.003 (3) | 0.003 (2) | 0.000 (3) |
| C10 | 0.032 (3) | 0.032 (4) | 0.025 (3) | 0.005 (3) | 0.007 (3) | −0.005 (3) |
| C11 | 0.035 (3) | 0.036 (4) | 0.029 (3) | 0.004 (3) | 0.003 (3) | −0.013 (3) |
| N12 | 0.024 (3) | 0.029 (4) | 0.029 (4) | 0.000 | 0.000 | −0.008 (3) |
| N13 | 0.028 (2) | 0.025 (3) | 0.025 (2) | −0.002 (2) | 0.004 (2) | −0.011 (2) |
| N14 | 0.027 (3) | 0.026 (4) | 0.027 (3) | 0.000 | 0.000 | 0.002 (3) |
| C15 | 0.026 (3) | 0.035 (4) | 0.023 (3) | −0.006 (3) | −0.001 (2) | −0.004 (3) |
| C16 | 0.025 (3) | 0.035 (4) | 0.038 (3) | −0.001 (3) | −0.002 (3) | −0.004 (3) |
| C17 | 0.034 (3) | 0.038 (5) | 0.049 (4) | −0.008 (3) | 0.001 (3) | −0.010 (3) |
| C18 | 0.029 (3) | 0.050 (5) | 0.029 (3) | −0.002 (3) | 0.001 (3) | −0.012 (3) |
| C19 | 0.042 (4) | 0.053 (5) | 0.036 (4) | −0.004 (4) | −0.008 (3) | 0.007 (3) |
| C20 | 0.037 (3) | 0.036 (4) | 0.041 (4) | −0.008 (3) | −0.009 (3) | 0.006 (3) |
| C21 | 0.027 (3) | 0.054 (5) | 0.037 (4) | −0.006 (3) | −0.001 (3) | −0.011 (3) |
| O22 | 0.047 (3) | 0.089 (5) | 0.136 (6) | 0.005 (3) | −0.027 (3) | −0.080 (5) |
| O23 | 0.046 (3) | 0.139 (6) | 0.036 (3) | −0.028 (3) | −0.016 (2) | 0.004 (3) |
| C24 | 0.023 (3) | 0.032 (4) | 0.034 (3) | −0.001 (3) | 0.003 (3) | −0.005 (3) |
| C25 | 0.049 (4) | 0.045 (4) | 0.045 (4) | 0.004 (4) | 0.011 (3) | 0.004 (4) |
| C26 | 0.044 (4) | 0.060 (5) | 0.034 (4) | −0.009 (4) | 0.020 (3) | −0.003 (3) |
| C27 | 0.028 (3) | 0.041 (5) | 0.046 (4) | 0.002 (3) | 0.001 (3) | −0.008 (3) |
| C28 | 0.042 (4) | 0.043 (5) | 0.048 (4) | 0.006 (3) | 0.005 (3) | −0.002 (3) |
| C29 | 0.034 (3) | 0.048 (5) | 0.035 (3) | 0.003 (3) | 0.009 (3) | −0.011 (3) |
| C30 | 0.035 (3) | 0.044 (5) | 0.039 (4) | −0.001 (3) | −0.001 (3) | −0.008 (3) |
| O31 | 0.035 (2) | 0.066 (4) | 0.052 (3) | −0.002 (3) | 0.014 (2) | −0.011 (3) |
| O32 | 0.043 (3) | 0.045 (4) | 0.055 (3) | 0.005 (2) | 0.004 (2) | −0.011 (2) |
| C33 | 0.065 (7) | 0.038 (8) | 0.129 (11) | 0.000 | −0.022 (7) | 0.000 |
| O34 | 0.113 (8) | 0.049 (7) | 0.211 (12) | 0.000 | −0.104 (8) | 0.000 |
Geometric parameters (Å, º) top
| Pd1—N14 | 2.004 (7) | C17—C18 | 1.398 (9) |
| Pd1—N12 | 2.015 (6) | C17—H17 | 0.9500 |
| Pd1—N13 | 2.018 (4) | C18—C19 | 1.386 (10) |
| Pd1—N13i | 2.018 (4) | C18—C21 | 1.515 (9) |
| C2—C2i | 1.359 (11) | C19—C20 | 1.391 (9) |
| C2—C3 | 1.442 (8) | C19—H19 | 0.9500 |
| C2—H2 | 0.9500 | C20—H20 | 0.9500 |
| C3—N12 | 1.377 (6) | C21—O22 | 1.230 (9) |
| C3—C4 | 1.398 (7) | C21—O23 | 1.272 (8) |
| C4—C5 | 1.402 (7) | O22—H22 | 0.8506 |
| C4—C15 | 1.489 (7) | O23—H23 | 0.8359 |
| C5—N13 | 1.368 (7) | C24—C25 | 1.390 (9) |
| C5—C6 | 1.438 (8) | C24—C29 | 1.396 (9) |
| C6—C7 | 1.359 (8) | C25—C26 | 1.405 (9) |
| C6—H6 | 0.9500 | C25—H25 | 0.9500 |
| C7—C8 | 1.442 (8) | C26—C27 | 1.370 (10) |
| C7—H7 | 0.9500 | C26—H26 | 0.9500 |
| C8—C9 | 1.389 (8) | C27—C28 | 1.367 (9) |
| C8—N13 | 1.390 (7) | C27—C30 | 1.528 (9) |
| C9—C10 | 1.402 (8) | C28—C29 | 1.410 (8) |
| C9—C24 | 1.485 (8) | C28—H28 | 0.9500 |
| C10—N14 | 1.385 (7) | C29—H29 | 0.9500 |
| C10—C11 | 1.446 (8) | C30—O32 | 1.238 (8) |
| C11—C11i | 1.372 (11) | C30—O31 | 1.274 (8) |
| C11—H11 | 0.9500 | O31—H31 | 0.8945 |
| N12—C3i | 1.377 (6) | C33—O34 | 1.378 (14) |
| N14—C10i | 1.385 (6) | C33—H33A | 0.9800 |
| C15—C16 | 1.396 (9) | C33—H33B | 0.9800 |
| C15—C20 | 1.403 (8) | C33—H33C | 0.9800 |
| C16—C17 | 1.395 (8) | O34—H34 | 0.8984 |
| C16—H16 | 0.9500 | | |
| | | |
| N14—Pd1—N12 | 179.1 (3) | C15—C16—H16 | 119.7 |
| N14—Pd1—N13 | 90.30 (13) | C17—C16—H16 | 119.7 |
| N12—Pd1—N13 | 89.73 (13) | C16—C17—C18 | 119.2 (7) |
| N14—Pd1—N13i | 90.30 (13) | C16—C17—H17 | 120.4 |
| N12—Pd1—N13i | 89.73 (13) | C18—C17—H17 | 120.4 |
| N13—Pd1—N13i | 175.5 (3) | C19—C18—C17 | 120.9 (6) |
| C2i—C2—C3 | 107.0 (3) | C19—C18—C21 | 120.1 (6) |
| C2i—C2—H2 | 126.5 | C17—C18—C21 | 118.9 (7) |
| C3—C2—H2 | 126.5 | C18—C19—C20 | 119.4 (6) |
| N12—C3—C4 | 125.6 (5) | C18—C19—H19 | 120.3 |
| N12—C3—C2 | 109.7 (5) | C20—C19—H19 | 120.3 |
| C4—C3—C2 | 124.5 (5) | C19—C20—C15 | 120.7 (7) |
| C3—C4—C5 | 123.7 (5) | C19—C20—H20 | 119.6 |
| C3—C4—C15 | 117.1 (5) | C15—C20—H20 | 119.6 |
| C5—C4—C15 | 119.2 (5) | O22—C21—O23 | 125.5 (7) |
| N13—C5—C4 | 125.6 (5) | O22—C21—C18 | 118.8 (7) |
| N13—C5—C6 | 110.3 (5) | O23—C21—C18 | 115.7 (7) |
| C4—C5—C6 | 124.0 (5) | C21—O22—H22 | 123.1 |
| C7—C6—C5 | 106.8 (5) | C21—O23—H23 | 117.0 |
| C7—C6—H6 | 126.6 | C25—C24—C29 | 118.5 (6) |
| C5—C6—H6 | 126.6 | C25—C24—C9 | 121.9 (6) |
| C6—C7—C8 | 107.4 (5) | C29—C24—C9 | 119.6 (5) |
| C6—C7—H7 | 126.3 | C24—C25—C26 | 120.0 (7) |
| C8—C7—H7 | 126.3 | C24—C25—H25 | 120.0 |
| C9—C8—N13 | 126.4 (5) | C26—C25—H25 | 120.0 |
| C9—C8—C7 | 124.4 (5) | C27—C26—C25 | 121.1 (7) |
| N13—C8—C7 | 109.1 (5) | C27—C26—H26 | 119.5 |
| C8—C9—C10 | 123.5 (5) | C25—C26—H26 | 119.5 |
| C8—C9—C24 | 117.9 (5) | C28—C27—C26 | 119.8 (6) |
| C10—C9—C24 | 118.6 (5) | C28—C27—C30 | 119.3 (7) |
| N14—C10—C9 | 126.0 (5) | C26—C27—C30 | 120.9 (6) |
| N14—C10—C11 | 110.2 (5) | C27—C28—C29 | 120.2 (7) |
| C9—C10—C11 | 123.7 (5) | C27—C28—H28 | 119.9 |
| C11i—C11—C10 | 106.8 (3) | C29—C28—H28 | 119.9 |
| C11i—C11—H11 | 126.6 | C24—C29—C28 | 120.5 (6) |
| C10—C11—H11 | 126.6 | C24—C29—H29 | 119.7 |
| C3i—N12—C3 | 106.3 (6) | C28—C29—H29 | 119.7 |
| C3i—N12—Pd1 | 126.6 (3) | O32—C30—O31 | 125.6 (6) |
| C3—N12—Pd1 | 126.6 (3) | O32—C30—C27 | 120.2 (6) |
| C5—N13—C8 | 106.3 (4) | O31—C30—C27 | 114.1 (7) |
| C5—N13—Pd1 | 127.1 (4) | C30—O31—H31 | 111.0 |
| C8—N13—Pd1 | 126.1 (4) | O34—C33—H33A | 109.5 |
| C10—N14—C10i | 105.8 (6) | O34—C33—H33B | 109.5 |
| C10—N14—Pd1 | 127.1 (3) | H33A—C33—H33B | 109.5 |
| C10i—N14—Pd1 | 127.1 (3) | O34—C33—H33C | 109.5 |
| C16—C15—C20 | 119.0 (5) | H33A—C33—H33C | 109.5 |
| C16—C15—C4 | 120.8 (5) | H33B—C33—H33C | 109.5 |
| C20—C15—C4 | 119.9 (6) | C33—O34—H34 | 120.8 |
| C15—C16—C17 | 120.6 (6) | | |
| Symmetry code: (i) −x, y, z. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O23—H23···O23ii | 0.84 | 1.84 | 2.542 (9) | 141 |
| O31—H31···O32iii | 0.89 | 1.77 | 2.669 (6) | 180 |
| O34—H34···O22iv | 0.90 | 1.71 | 2.608 (7) | 179 |
| Symmetry codes: (ii) −x+1/2, y, −z+3/2; (iii) −x+1/2, −y−1/2, −z; (iv) −x+1/2, −y+1/2, −z+1. |
(II) [5,10,15,20-tetrakis(4-carboxyphenyl)porphyrinato]palladium(II)–
N,
N'-dimethylformamide–water (1/4/4)
top
Crystal data top
| [Pd(C48H28N4O8)]·4C3H7NO·4H2O | Z = 1 |
| Mr = 1259.61 | F(000) = 654 |
| Triclinic, P1 | Dx = 1.418 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 7.7243 (2) Å | Cell parameters from 5044 reflections |
| b = 13.2668 (4) Å | θ = 1.4–25.7° |
| c = 14.5878 (5) Å | µ = 0.39 mm−1 |
| α = 81.3820 (16)° | T = 110 K |
| β = 87.3282 (17)° | Needles, red |
| γ = 87.0384 (17)° | 0.60 × 0.10 × 0.10 mm |
| V = 1474.91 (8) Å3 | |
Data collection top
Nonius KappaCCD
diffractometer | 4868 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.045 |
| Graphite monochromator | θmax = 25.7°, θmin = 2.3° |
| Detector resolution: 12.8 pixels mm-1 | h = −9→9 |
| 1 deg. ϕ and ω scans | k = −15→16 |
| 12855 measured reflections | l = −17→17 |
| 5498 independent reflections | |
Refinement top
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full with fixed elements per cycle | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.083 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0221P)2 + 1.3991P]
where P = (Fo2 + 2Fc2)/3 |
| 5498 reflections | (Δ/σ)max = 0.049 |
| 437 parameters | Δρmax = 0.31 e Å−3 |
| 8 restraints | Δρmin = −0.72 e Å−3 |
Crystal data top
| [Pd(C48H28N4O8)]·4C3H7NO·4H2O | γ = 87.0384 (17)° |
| Mr = 1259.61 | V = 1474.91 (8) Å3 |
| Triclinic, P1 | Z = 1 |
| a = 7.7243 (2) Å | Mo Kα radiation |
| b = 13.2668 (4) Å | µ = 0.39 mm−1 |
| c = 14.5878 (5) Å | T = 110 K |
| α = 81.3820 (16)° | 0.60 × 0.10 × 0.10 mm |
| β = 87.3282 (17)° | |
Data collection top
Nonius KappaCCD
diffractometer | 4868 reflections with I > 2σ(I) |
| 12855 measured reflections | Rint = 0.045 |
| 5498 independent reflections | |
Refinement top
| R[F2 > 2σ(F2)] = 0.037 | 8 restraints |
| wR(F2) = 0.083 | H-atom parameters constrained |
| S = 1.02 | Δρmax = 0.31 e Å−3 |
| 5498 reflections | Δρmin = −0.72 e Å−3 |
| 437 parameters | |
Special details top
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes)
are estimated using the full covariance matrix. The cell e.s.d.'s are taken
into account individually in the estimation of e.s.d.'s in distances, angles
and torsion angles; correlations between e.s.d.'s in cell parameters are only
used when they are defined by crystal symmetry. An approximate (isotropic)
treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s.
planes. |
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor
wR and goodness of fit S are based on F2, conventional
R-factors R are based on F, with F set to zero for
negative F2. The threshold expression of F2 >
σ(F2) is used only for calculating R-factors(gt) etc.
and is not relevant to the choice of reflections for refinement.
R-factors based on F2 are statistically about twice as large
as those based on F, and R- factors based on ALL data will be
even larger. One of the DMF solvent molecules is orientationally disordered between two
sites. The corresponding species are represented by atoms O39A through C43A
and O39B through C43B, respectively. The bond distances in these disordered
species were restrained to normal values to avoid irregular geometries. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2) top| | x | y | z | Uiso*/Ueq | Occ. (<1) |
| Pd1 | 0.0000 | 0.0000 | 1.0000 | 0.01444 (8) | |
| C2 | −0.2902 (3) | −0.13339 (18) | 0.96293 (17) | 0.0157 (5) | |
| C3 | −0.4005 (3) | −0.15028 (18) | 0.89005 (17) | 0.0178 (5) | |
| H3 | −0.4784 | −0.2038 | 0.8925 | 0.021* | |
| C4 | −0.3726 (3) | −0.07618 (18) | 0.81786 (17) | 0.0185 (5) | |
| H4 | −0.4269 | −0.0680 | 0.7599 | 0.022* | |
| C5 | −0.2451 (3) | −0.01155 (17) | 0.84459 (16) | 0.0138 (5) | |
| C6 | −0.1837 (3) | 0.07443 (18) | 0.78745 (16) | 0.0151 (5) | |
| C7 | −0.0485 (3) | 0.13108 (17) | 0.80966 (16) | 0.0147 (5) | |
| C8 | 0.0216 (3) | 0.21695 (18) | 0.75038 (17) | 0.0163 (5) | |
| H8 | −0.0169 | 0.2454 | 0.6907 | 0.020* | |
| C9 | 0.1524 (3) | 0.24978 (18) | 0.79508 (16) | 0.0157 (5) | |
| H9 | 0.2238 | 0.3050 | 0.7723 | 0.019* | |
| C10 | 0.1635 (3) | 0.18551 (18) | 0.88381 (16) | 0.0150 (5) | |
| C11 | 0.2818 (3) | 0.19532 (17) | 0.95141 (17) | 0.0151 (5) | |
| N12 | −0.1937 (2) | −0.04911 (14) | 0.93278 (13) | 0.0148 (4) | |
| N13 | 0.0397 (2) | 0.11294 (14) | 0.89106 (13) | 0.0132 (4) | |
| C14 | −0.2772 (3) | 0.10618 (17) | 0.69914 (16) | 0.0155 (5) | |
| C15 | −0.4538 (3) | 0.13467 (19) | 0.70647 (17) | 0.0206 (5) | |
| H15 | −0.5057 | 0.1424 | 0.7655 | 0.025* | |
| C16 | −0.5538 (3) | 0.1517 (2) | 0.62900 (18) | 0.0216 (5) | |
| H16 | −0.6738 | 0.1700 | 0.6352 | 0.026* | |
| C17 | −0.4786 (3) | 0.14203 (18) | 0.54186 (17) | 0.0177 (5) | |
| C18 | −0.3008 (3) | 0.11919 (18) | 0.53311 (18) | 0.0199 (5) | |
| H18 | −0.2476 | 0.1164 | 0.4734 | 0.024* | |
| C19 | −0.2012 (3) | 0.10041 (18) | 0.61135 (17) | 0.0180 (5) | |
| H19 | −0.0806 | 0.0835 | 0.6050 | 0.022* | |
| C20 | −0.5879 (3) | 0.15329 (18) | 0.45888 (18) | 0.0206 (5) | |
| O21 | −0.5271 (2) | 0.15510 (14) | 0.37971 (12) | 0.0257 (4) | |
| O22 | −0.7552 (2) | 0.15908 (16) | 0.47920 (13) | 0.0335 (5) | |
| H22 | −0.8358 | 0.1584 | 0.4323 | 0.040* | |
| C23 | 0.4094 (3) | 0.27708 (18) | 0.93129 (16) | 0.0158 (5) | |
| C24 | 0.3565 (3) | 0.38050 (18) | 0.91880 (17) | 0.0182 (5) | |
| H24 | 0.2368 | 0.3998 | 0.9245 | 0.022* | |
| C25 | 0.4772 (3) | 0.45474 (18) | 0.89815 (17) | 0.0192 (5) | |
| H25 | 0.4398 | 0.5247 | 0.8899 | 0.023* | |
| C26 | 0.6540 (3) | 0.42765 (18) | 0.88928 (17) | 0.0170 (5) | |
| C27 | 0.7077 (3) | 0.32493 (18) | 0.90345 (17) | 0.0165 (5) | |
| H27 | 0.8277 | 0.3058 | 0.8987 | 0.020* | |
| C28 | 0.5871 (3) | 0.25063 (18) | 0.92442 (17) | 0.0167 (5) | |
| H28 | 0.6251 | 0.1807 | 0.9343 | 0.020* | |
| C29 | 0.7804 (3) | 0.50806 (19) | 0.85813 (18) | 0.0200 (5) | |
| O30 | 0.9438 (2) | 0.47405 (13) | 0.85937 (14) | 0.0265 (4) | |
| H30 | 1.0188 | 0.5245 | 0.8220 | 0.032* | |
| O31 | 0.7367 (2) | 0.59738 (13) | 0.83154 (14) | 0.0283 (4) | |
| O32 | 0.8576 (2) | 0.42381 (14) | 0.25966 (14) | 0.0307 (4) | |
| H32A | 0.8245 | 0.4613 | 0.3052 | 0.037* | |
| H32B | 0.7551 | 0.3860 | 0.2548 | 0.037* | |
| O33 | 0.5663 (2) | 0.32050 (14) | 0.25003 (15) | 0.0343 (5) | |
| H33A | 0.5382 | 0.2612 | 0.2870 | 0.041* | |
| H33B | 0.4639 | 0.3520 | 0.2323 | 0.041* | |
| O34 | 1.0397 (2) | 0.14458 (14) | 0.35328 (12) | 0.0263 (4) | |
| C35 | 1.0357 (3) | 0.19679 (19) | 0.27469 (18) | 0.0206 (5) | |
| H35 | 1.1119 | 0.2515 | 0.2605 | 0.025* | |
| N36 | 0.9309 (3) | 0.17978 (15) | 0.21023 (14) | 0.0191 (4) | |
| C37 | 0.9205 (3) | 0.2473 (2) | 0.12174 (18) | 0.0255 (6) | |
| H37A | 0.8085 | 0.2858 | 0.1194 | 0.038* | |
| H37B | 0.9313 | 0.2066 | 0.0708 | 0.038* | |
| H37C | 1.0146 | 0.2950 | 0.1156 | 0.038* | |
| C38 | 0.8107 (3) | 0.0975 (2) | 0.2296 (2) | 0.0246 (6) | |
| H38A | 0.8556 | 0.0454 | 0.2787 | 0.037* | |
| H38B | 0.7983 | 0.0669 | 0.1733 | 0.037* | |
| H38C | 0.6974 | 0.1248 | 0.2499 | 0.037* | |
| O39A | 0.1977 (8) | 0.4360 (4) | 0.6152 (3) | 0.0574 (17) | 0.460 (4) |
| C40A | 0.2154 (10) | 0.4309 (4) | 0.5312 (3) | 0.0418 (18) | 0.460 (4) |
| H40A | 0.2039 | 0.3661 | 0.5124 | 0.050* | 0.460 (4) |
| N41A | 0.2497 (6) | 0.5099 (3) | 0.4642 (3) | 0.0285 (13) | 0.460 (4) |
| C42A | 0.2842 (11) | 0.6100 (4) | 0.4874 (5) | 0.050 (2) | 0.460 (4) |
| H42A | 0.1749 | 0.6506 | 0.4903 | 0.074* | 0.460 (4) |
| H42B | 0.3627 | 0.6450 | 0.4397 | 0.074* | 0.460 (4) |
| H42C | 0.3380 | 0.6018 | 0.5477 | 0.074* | 0.460 (4) |
| C43A | 0.2634 (12) | 0.4953 (7) | 0.3669 (3) | 0.056 (2) | 0.460 (4) |
| H43A | 0.2463 | 0.4236 | 0.3622 | 0.083* | 0.460 (4) |
| H43B | 0.3785 | 0.5139 | 0.3408 | 0.083* | 0.460 (4) |
| H43C | 0.1744 | 0.5385 | 0.3324 | 0.083* | 0.460 (4) |
| O39B | 0.2072 (7) | 0.4754 (3) | 0.5985 (3) | 0.0615 (15) | 0.540 (4) |
| C40B | 0.2490 (8) | 0.5360 (4) | 0.5288 (3) | 0.0508 (18) | 0.540 (4) |
| H40B | 0.2614 | 0.6052 | 0.5363 | 0.061* | 0.540 (4) |
| N41B | 0.2782 (6) | 0.5108 (3) | 0.4432 (3) | 0.0421 (14) | 0.540 (4) |
| C42B | 0.2411 (10) | 0.4087 (4) | 0.4259 (5) | 0.059 (2) | 0.540 (4) |
| H42D | 0.2022 | 0.3678 | 0.4841 | 0.089* | 0.540 (4) |
| H42E | 0.3464 | 0.3762 | 0.4013 | 0.089* | 0.540 (4) |
| H42F | 0.1499 | 0.4136 | 0.3808 | 0.089* | 0.540 (4) |
| C43B | 0.3229 (10) | 0.5875 (5) | 0.3643 (4) | 0.058 (2) | 0.540 (4) |
| H43D | 0.3278 | 0.6541 | 0.3852 | 0.086* | 0.540 (4) |
| H43E | 0.2347 | 0.5912 | 0.3176 | 0.086* | 0.540 (4) |
| H43F | 0.4363 | 0.5687 | 0.3371 | 0.086* | 0.540 (4) |
Atomic displacement parameters (Å2) top| | U11 | U22 | U33 | U12 | U13 | U23 |
| Pd1 | 0.01361 (14) | 0.01625 (14) | 0.01373 (15) | −0.00118 (10) | −0.00450 (10) | −0.00173 (10) |
| C2 | 0.0125 (11) | 0.0180 (12) | 0.0172 (13) | −0.0010 (10) | −0.0021 (10) | −0.0035 (10) |
| C3 | 0.0148 (11) | 0.0214 (12) | 0.0178 (13) | −0.0043 (10) | −0.0055 (10) | −0.0025 (10) |
| C4 | 0.0160 (12) | 0.0226 (13) | 0.0177 (13) | −0.0027 (10) | −0.0068 (10) | −0.0032 (10) |
| C5 | 0.0107 (10) | 0.0182 (12) | 0.0132 (12) | −0.0002 (9) | −0.0028 (9) | −0.0035 (9) |
| C6 | 0.0128 (11) | 0.0201 (12) | 0.0128 (12) | 0.0008 (10) | −0.0031 (9) | −0.0034 (9) |
| C7 | 0.0121 (11) | 0.0174 (12) | 0.0144 (12) | 0.0015 (9) | −0.0010 (9) | −0.0023 (9) |
| C8 | 0.0153 (11) | 0.0184 (12) | 0.0148 (12) | 0.0006 (10) | −0.0039 (10) | −0.0003 (10) |
| C9 | 0.0130 (11) | 0.0181 (12) | 0.0160 (12) | −0.0019 (10) | −0.0021 (9) | −0.0015 (10) |
| C10 | 0.0136 (11) | 0.0179 (12) | 0.0139 (12) | −0.0007 (10) | 0.0002 (9) | −0.0039 (9) |
| C11 | 0.0129 (11) | 0.0153 (11) | 0.0178 (13) | 0.0011 (9) | −0.0041 (10) | −0.0043 (9) |
| N12 | 0.0151 (9) | 0.0156 (10) | 0.0136 (10) | 0.0008 (8) | −0.0031 (8) | −0.0019 (8) |
| N13 | 0.0127 (9) | 0.0155 (10) | 0.0119 (10) | −0.0005 (8) | −0.0031 (8) | −0.0027 (8) |
| C14 | 0.0172 (11) | 0.0141 (11) | 0.0155 (12) | −0.0009 (10) | −0.0043 (10) | −0.0020 (9) |
| C15 | 0.0176 (12) | 0.0296 (14) | 0.0145 (13) | 0.0021 (11) | −0.0014 (10) | −0.0042 (11) |
| C16 | 0.0148 (12) | 0.0299 (14) | 0.0203 (13) | 0.0039 (11) | −0.0047 (10) | −0.0044 (11) |
| C17 | 0.0175 (12) | 0.0192 (12) | 0.0161 (13) | −0.0010 (10) | −0.0066 (10) | 0.0003 (10) |
| C18 | 0.0199 (12) | 0.0232 (13) | 0.0161 (13) | 0.0010 (11) | −0.0016 (10) | −0.0017 (10) |
| C19 | 0.0139 (11) | 0.0207 (12) | 0.0193 (13) | −0.0004 (10) | −0.0024 (10) | −0.0021 (10) |
| C20 | 0.0230 (13) | 0.0207 (13) | 0.0183 (14) | −0.0008 (11) | −0.0060 (11) | −0.0026 (10) |
| O21 | 0.0280 (10) | 0.0348 (10) | 0.0143 (10) | −0.0031 (8) | −0.0066 (8) | −0.0012 (8) |
| O22 | 0.0182 (9) | 0.0632 (14) | 0.0209 (10) | 0.0054 (9) | −0.0107 (8) | −0.0114 (9) |
| C23 | 0.0167 (11) | 0.0189 (12) | 0.0124 (12) | −0.0030 (10) | −0.0027 (9) | −0.0032 (9) |
| C24 | 0.0137 (11) | 0.0204 (12) | 0.0208 (13) | −0.0012 (10) | −0.0037 (10) | −0.0029 (10) |
| C25 | 0.0186 (12) | 0.0165 (12) | 0.0220 (14) | 0.0001 (10) | −0.0028 (10) | −0.0011 (10) |
| C26 | 0.0163 (12) | 0.0196 (12) | 0.0157 (12) | −0.0036 (10) | −0.0031 (10) | −0.0031 (10) |
| C27 | 0.0113 (11) | 0.0206 (12) | 0.0176 (13) | 0.0010 (10) | −0.0028 (9) | −0.0027 (10) |
| C28 | 0.0158 (11) | 0.0160 (12) | 0.0184 (13) | −0.0017 (10) | −0.0023 (10) | −0.0014 (10) |
| C29 | 0.0167 (12) | 0.0228 (13) | 0.0214 (14) | −0.0023 (10) | −0.0033 (10) | −0.0046 (11) |
| O30 | 0.0142 (8) | 0.0218 (9) | 0.0412 (12) | −0.0041 (7) | −0.0015 (8) | 0.0038 (8) |
| O31 | 0.0202 (9) | 0.0183 (9) | 0.0446 (12) | −0.0030 (8) | −0.0003 (9) | 0.0017 (8) |
| O32 | 0.0243 (10) | 0.0319 (10) | 0.0369 (12) | −0.0093 (8) | 0.0050 (9) | −0.0074 (9) |
| O33 | 0.0214 (9) | 0.0251 (10) | 0.0524 (14) | −0.0019 (8) | −0.0012 (9) | 0.0081 (9) |
| O34 | 0.0232 (9) | 0.0346 (10) | 0.0210 (10) | −0.0015 (8) | −0.0104 (8) | −0.0010 (8) |
| C35 | 0.0133 (11) | 0.0248 (13) | 0.0248 (14) | 0.0010 (10) | −0.0066 (10) | −0.0062 (11) |
| N36 | 0.0160 (10) | 0.0215 (11) | 0.0201 (11) | 0.0018 (9) | −0.0051 (9) | −0.0033 (9) |
| C37 | 0.0256 (13) | 0.0294 (14) | 0.0210 (14) | 0.0046 (12) | −0.0055 (11) | −0.0027 (11) |
| C38 | 0.0183 (12) | 0.0260 (14) | 0.0314 (16) | −0.0009 (11) | −0.0051 (11) | −0.0089 (12) |
| O39A | 0.102 (5) | 0.032 (3) | 0.036 (3) | −0.006 (3) | 0.008 (3) | 0.001 (2) |
| C40A | 0.059 (5) | 0.032 (4) | 0.037 (4) | −0.006 (3) | 0.002 (3) | −0.013 (3) |
| N41A | 0.034 (3) | 0.027 (3) | 0.024 (3) | 0.001 (2) | 0.005 (2) | −0.006 (2) |
| C42A | 0.086 (6) | 0.028 (4) | 0.034 (4) | −0.017 (4) | −0.002 (4) | 0.006 (3) |
| C43A | 0.076 (6) | 0.068 (5) | 0.024 (4) | 0.004 (5) | −0.001 (4) | −0.010 (4) |
| O39B | 0.095 (4) | 0.062 (3) | 0.030 (3) | −0.022 (3) | 0.018 (2) | −0.016 (2) |
| C40B | 0.055 (4) | 0.048 (4) | 0.057 (5) | 0.005 (3) | −0.009 (3) | −0.030 (3) |
| N41B | 0.044 (3) | 0.047 (3) | 0.037 (3) | 0.011 (2) | 0.002 (2) | −0.017 (2) |
| C42B | 0.088 (5) | 0.046 (4) | 0.048 (4) | 0.009 (4) | −0.005 (4) | −0.023 (3) |
| C43B | 0.076 (5) | 0.051 (4) | 0.042 (4) | 0.008 (4) | 0.001 (4) | 0.001 (3) |
Geometric parameters (Å, º) top
| Pd1—N12 | 2.0133 (19) | C26—C27 | 1.392 (3) |
| Pd1—N12i | 2.0133 (19) | C26—C29 | 1.490 (3) |
| Pd1—N13 | 2.0379 (19) | C27—C28 | 1.383 (3) |
| Pd1—N13i | 2.0379 (19) | C27—H27 | 0.9500 |
| C2—N12 | 1.381 (3) | C28—H28 | 0.9500 |
| C2—C11i | 1.391 (3) | C29—O31 | 1.225 (3) |
| C2—C3 | 1.443 (3) | C29—O30 | 1.318 (3) |
| C3—C4 | 1.346 (3) | O30—H30 | 0.9905 |
| C3—H3 | 0.9500 | O32—H32A | 0.9084 |
| C4—C5 | 1.442 (3) | O32—H32B | 0.9694 |
| C4—H4 | 0.9500 | O33—H33A | 0.9147 |
| C5—N12 | 1.377 (3) | O33—H33B | 0.9056 |
| C5—C6 | 1.398 (3) | O34—C35 | 1.248 (3) |
| C6—C7 | 1.395 (3) | C35—N36 | 1.321 (3) |
| C6—C14 | 1.500 (3) | C35—H35 | 0.9500 |
| C7—N13 | 1.380 (3) | N36—C38 | 1.456 (3) |
| C7—C8 | 1.437 (3) | N36—C37 | 1.460 (3) |
| C8—C9 | 1.352 (3) | C37—H37A | 0.9800 |
| C8—H8 | 0.9500 | C37—H37B | 0.9800 |
| C9—C10 | 1.443 (3) | C37—H37C | 0.9800 |
| C9—H9 | 0.9500 | C38—H38A | 0.9800 |
| C10—N13 | 1.381 (3) | C38—H38B | 0.9800 |
| C10—C11 | 1.400 (3) | C38—H38C | 0.9800 |
| C11—C2i | 1.391 (3) | O39A—C40A | 1.239 (3) |
| C11—C23 | 1.491 (3) | C40A—N41A | 1.349 (3) |
| C14—C19 | 1.395 (3) | C40A—H40A | 0.9500 |
| C14—C15 | 1.400 (3) | N41A—C42A | 1.460 (3) |
| C15—C16 | 1.382 (3) | N41A—C43A | 1.460 (3) |
| C15—H15 | 0.9500 | C42A—H42A | 0.9800 |
| C16—C17 | 1.394 (4) | C42A—H42B | 0.9800 |
| C16—H16 | 0.9500 | C42A—H42C | 0.9800 |
| C17—C18 | 1.395 (3) | C43A—H43A | 0.9800 |
| C17—C20 | 1.493 (3) | C43A—H43B | 0.9800 |
| C18—C19 | 1.391 (3) | C43A—H43C | 0.9800 |
| C18—H18 | 0.9500 | O39B—C40B | 1.240 (3) |
| C19—H19 | 0.9500 | C40B—N41B | 1.347 (3) |
| C20—O21 | 1.224 (3) | C40B—H40B | 0.9500 |
| C20—O22 | 1.313 (3) | N41B—C42B | 1.458 (3) |
| O22—H22 | 0.9473 | N41B—C43B | 1.459 (3) |
| C23—C24 | 1.399 (3) | C42B—H42D | 0.9800 |
| C23—C28 | 1.401 (3) | C42B—H42E | 0.9800 |
| C24—C25 | 1.382 (3) | C42B—H42F | 0.9800 |
| C24—H24 | 0.9500 | C43B—H43D | 0.9800 |
| C25—C26 | 1.399 (3) | C43B—H43E | 0.9800 |
| C25—H25 | 0.9500 | C43B—H43F | 0.9800 |
| | | |
| N12—Pd1—N12i | 180.0 | C24—C23—C11 | 121.6 (2) |
| N12—Pd1—N13 | 89.59 (8) | C28—C23—C11 | 119.7 (2) |
| N12i—Pd1—N13 | 90.41 (8) | C25—C24—C23 | 120.4 (2) |
| N12—Pd1—N13i | 90.41 (8) | C25—C24—H24 | 119.8 |
| N12i—Pd1—N13i | 89.59 (8) | C23—C24—H24 | 119.8 |
| N13—Pd1—N13i | 180.0 | C24—C25—C26 | 120.5 (2) |
| N12—C2—C11i | 126.5 (2) | C24—C25—H25 | 119.7 |
| N12—C2—C3 | 109.3 (2) | C26—C25—H25 | 119.7 |
| C11i—C2—C3 | 124.2 (2) | C27—C26—C25 | 119.3 (2) |
| C4—C3—C2 | 107.4 (2) | C27—C26—C29 | 120.8 (2) |
| C4—C3—H3 | 126.3 | C25—C26—C29 | 119.8 (2) |
| C2—C3—H3 | 126.3 | C28—C27—C26 | 120.2 (2) |
| C3—C4—C5 | 107.4 (2) | C28—C27—H27 | 119.9 |
| C3—C4—H4 | 126.3 | C26—C27—H27 | 119.9 |
| C5—C4—H4 | 126.3 | C27—C28—C23 | 120.9 (2) |
| N12—C5—C6 | 126.5 (2) | C27—C28—H28 | 119.6 |
| N12—C5—C4 | 109.4 (2) | C23—C28—H28 | 119.6 |
| C6—C5—C4 | 124.0 (2) | O31—C29—O30 | 122.8 (2) |
| C7—C6—C5 | 124.0 (2) | O31—C29—C26 | 123.2 (2) |
| C7—C6—C14 | 120.4 (2) | O30—C29—C26 | 114.0 (2) |
| C5—C6—C14 | 115.6 (2) | C29—O30—H30 | 110.5 |
| N13—C7—C6 | 125.5 (2) | H32A—O32—H32B | 102.2 |
| N13—C7—C8 | 109.5 (2) | H33A—O33—H33B | 105.5 |
| C6—C7—C8 | 125.0 (2) | O34—C35—N36 | 123.2 (2) |
| C9—C8—C7 | 107.4 (2) | O34—C35—H35 | 118.4 |
| C9—C8—H8 | 126.3 | N36—C35—H35 | 118.4 |
| C7—C8—H8 | 126.3 | C35—N36—C38 | 120.0 (2) |
| C8—C9—C10 | 107.3 (2) | C35—N36—C37 | 121.6 (2) |
| C8—C9—H9 | 126.3 | C38—N36—C37 | 118.2 (2) |
| C10—C9—H9 | 126.3 | N36—C37—H37A | 109.5 |
| N13—C10—C11 | 125.6 (2) | N36—C37—H37B | 109.5 |
| N13—C10—C9 | 109.23 (19) | H37A—C37—H37B | 109.5 |
| C11—C10—C9 | 125.1 (2) | N36—C37—H37C | 109.5 |
| C2i—C11—C10 | 124.6 (2) | H37A—C37—H37C | 109.5 |
| C2i—C11—C23 | 116.8 (2) | H37B—C37—H37C | 109.5 |
| C10—C11—C23 | 118.6 (2) | N36—C38—H38A | 109.5 |
| C5—N12—C2 | 106.44 (19) | N36—C38—H38B | 109.5 |
| C5—N12—Pd1 | 126.95 (16) | H38A—C38—H38B | 109.5 |
| C2—N12—Pd1 | 126.36 (15) | N36—C38—H38C | 109.5 |
| C7—N13—C10 | 106.49 (19) | H38A—C38—H38C | 109.5 |
| C7—N13—Pd1 | 127.17 (15) | H38B—C38—H38C | 109.5 |
| C10—N13—Pd1 | 126.33 (15) | O39A—C40A—N41A | 125.2 (5) |
| C19—C14—C15 | 118.8 (2) | O39A—C40A—H40A | 117.4 |
| C19—C14—C6 | 123.4 (2) | N41A—C40A—H40A | 117.4 |
| C15—C14—C6 | 117.6 (2) | C40A—N41A—C42A | 121.1 (5) |
| C16—C15—C14 | 120.9 (2) | C40A—N41A—C43A | 120.4 (5) |
| C16—C15—H15 | 119.5 | C42A—N41A—C43A | 118.4 (5) |
| C14—C15—H15 | 119.5 | O39B—C40B—N41B | 124.6 (5) |
| C15—C16—C17 | 120.1 (2) | O39B—C40B—H40B | 117.7 |
| C15—C16—H16 | 120.0 | N41B—C40B—H40B | 117.7 |
| C17—C16—H16 | 120.0 | C40B—N41B—C42B | 119.7 (5) |
| C16—C17—C18 | 119.5 (2) | C40B—N41B—C43B | 121.1 (5) |
| C16—C17—C20 | 120.3 (2) | C42B—N41B—C43B | 118.7 (5) |
| C18—C17—C20 | 120.2 (2) | N41B—C42B—H42D | 109.5 |
| C19—C18—C17 | 120.3 (2) | N41B—C42B—H42E | 109.5 |
| C19—C18—H18 | 119.9 | H42D—C42B—H42E | 109.5 |
| C17—C18—H18 | 119.9 | N41B—C42B—H42F | 109.5 |
| C18—C19—C14 | 120.4 (2) | H42D—C42B—H42F | 109.5 |
| C18—C19—H19 | 119.8 | H42E—C42B—H42F | 109.5 |
| C14—C19—H19 | 119.8 | N41B—C43B—H43D | 109.5 |
| O21—C20—O22 | 123.4 (2) | N41B—C43B—H43E | 109.5 |
| O21—C20—C17 | 123.0 (2) | H43D—C43B—H43E | 109.5 |
| O22—C20—C17 | 113.6 (2) | N41B—C43B—H43F | 109.5 |
| C20—O22—H22 | 120.1 | H43D—C43B—H43F | 109.5 |
| C24—C23—C28 | 118.7 (2) | H43E—C43B—H43F | 109.5 |
| Symmetry code: (i) −x, −y, −z+2. |
Hydrogen-bond geometry (Å, º) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O22—H22···O34ii | 0.95 | 1.57 | 2.516 (2) | 173 |
| O30—H30···O32iii | 0.99 | 1.59 | 2.543 (3) | 161 |
| O32—H32A···O39Biv | 0.91 | 1.74 | 2.642 (4) | 170 |
| O32—H32A···O39Aiv | 0.91 | 1.91 | 2.799 (5) | 164 |
| O32—H32B···O33 | 0.97 | 1.75 | 2.714 (3) | 177 |
| O33—H33A···O21v | 0.91 | 1.87 | 2.776 (3) | 169 |
| O33—H33B···O31iv | 0.91 | 1.89 | 2.773 (3) | 166 |
| Symmetry codes: (ii) x−2, y, z; (iii) −x+2, −y+1, −z+1; (iv) −x+1, −y+1, −z+1; (v) x+1, y, z. |
Experimental details
| | (I) | (II) |
| Crystal data |
| Chemical formula | [Pd(C48H28N4O8)]·CH4O | [Pd(C48H28N4O8)]·4C3H7NO·4H2O |
| Mr | 927.21 | 1259.61 |
| Crystal system, space group | Orthorhombic, Cmca | Triclinic, P1 |
| Temperature (K) | 110 | 110 |
| a, b, c (Å) | 31.0347 (6), 15.9441 (11), 18.3824 (7) | 7.7243 (2), 13.2668 (4), 14.5878 (5) |
| α, β, γ (°) | 90, 90, 90 | 81.3820 (16), 87.3282 (17), 87.0384 (17) |
| V (Å3) | 9096.0 (7) | 1474.91 (8) |
| Z | 8 | 1 |
| Radiation type | Mo Kα | Mo Kα |
| µ (mm−1) | 0.47 | 0.39 |
| Crystal size (mm) | 0.30 × 0.10 × 0.05 | 0.60 × 0.10 × 0.10 |
| |
| Data collection |
| Diffractometer | Nonius KappaCCD
diffractometer | Nonius KappaCCD
diffractometer |
| Absorption correction | – | – |
No. of measured, independent and
observed [I > 2σ(I)] reflections | 20365, 4344, 2688 | 12855, 5498, 4868 |
| Rint | 0.098 | 0.045 |
| (sin θ/λ)max (Å−1) | 0.608 | 0.609 |
| |
| Refinement |
| R[F2 > 2σ(F2)], wR(F2), S | 0.070, 0.160, 1.00 | 0.037, 0.083, 1.02 |
| No. of reflections | 4344 | 5498 |
| No. of parameters | 291 | 437 |
| No. of restraints | 12 | 8 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.50, −0.77 | 0.31, −0.72 |
Hydrogen-bond geometry (Å, º) for (I) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O23—H23···O23i | 0.84 | 1.84 | 2.542 (9) | 141 |
| O31—H31···O32ii | 0.89 | 1.77 | 2.669 (6) | 180 |
| O34—H34···O22iii | 0.90 | 1.71 | 2.608 (7) | 179 |
| Symmetry codes: (i) −x+1/2, y, −z+3/2; (ii) −x+1/2, −y−1/2, −z; (iii) −x+1/2, −y+1/2, −z+1. |
Hydrogen-bond geometry (Å, º) for (II) top
| D—H···A | D—H | H···A | D···A | D—H···A |
| O22—H22···O34i | 0.95 | 1.57 | 2.516 (2) | 173 |
| O30—H30···O32ii | 0.99 | 1.59 | 2.543 (3) | 161 |
| O32—H32A···O39Biii | 0.91 | 1.74 | 2.642 (4) | 170 |
| O32—H32A···O39Aiii | 0.91 | 1.91 | 2.799 (5) | 164 |
| O32—H32B···O33 | 0.97 | 1.75 | 2.714 (3) | 177 |
| O33—H33A···O21iv | 0.91 | 1.87 | 2.776 (3) | 169 |
| O33—H33B···O31iii | 0.91 | 1.89 | 2.773 (3) | 166 |
| Symmetry codes: (i) x−2, y, z; (ii) −x+2, −y+1, −z+1; (iii) −x+1, −y+1, −z+1; (iv) x+1, y, z. |

 journal menu
journal menu








![[Figure 1]](http://journals.iucr.org/c/issues/2008/01/00/gd3172/gd3172fig1thm.gif)
![[Figure 2]](http://journals.iucr.org/c/issues/2008/01/00/gd3172/gd3172fig2thm.gif)
![[Figure 3]](http://journals.iucr.org/c/issues/2008/01/00/gd3172/gd3172fig3thm.gif)
![[Figure 4]](http://journals.iucr.org/c/issues/2008/01/00/gd3172/gd3172fig4thm.gif)
![[Figure 5]](http://journals.iucr.org/c/issues/2008/01/00/gd3172/gd3172fig5thm.gif)
![[Figure 6]](http://journals.iucr.org/c/issues/2008/01/00/gd3172/gd3172fig6thm.gif)




Metalloporphyrin macrocycles when tetra-substituted with 4-carboxyphenyl groups at their meso positions (M-TCPP) provide a classical example of an organic species with multiple complementary terminal functional groups directed at four diverging directions of the equatorial molecular plane, which exhibit high a propensity for self-assembling into flat hydrogen-bonded two-dimensional nets (Goldberg, 2005, and references therein). It has been shown that the carboxylic acid functions may engage readily in two different supramolecular synthons, either catemeric (chain-type) or cyclic dimeric. In the latter case, utilization of four head-to-head (COOH)2 cyclic dimeric associations to four neighboring porphyrin units leads to the formation of quadrangular grid arrays with large pores (Dastidar et al., 1996; Diskin-Posner & Goldberg, 1999; George & Goldberg, 2006; George et al., 2006; Lipstman, Muniappan & Goldberg, 2007). In the solid, these pores are filled either by a guest template or by self-interpenetration of the networks. Every molecule of the tetra-acid within such supramolecular layered assembly is involved simultaneously in eight nearly linear O—H···O interactions, or four (COOH)2 R22(8)-type (Etter, 1990; Bernstein et al., 1995) synthons.
In non-interwoven structures the two-dimensional multiporphyrin layers form offset stacks along the normal direction, held together by van der Waals forces. In addition to supramolecular isomerism of the catenane type (occurrence of interwoven versus. non-interpenetrating networks), conformational isomerism, where arrays of identical composition reveal different grid shapes, has also been observed in these systems (George et al., 2006). The neat networking of the M-TCPP units into homogeneous multi-porphyrin nets can be readily disrupted by competing solvation that can modify or prevent supramolecular association. This is common with strong Lewis base reagents, e.g. dimethyl sulfoxide and pyridine, which each have a higher proton affinity than the carbonyl fragments of the carboxylic acid (Lipstman et al., 2006; George et al., 2006). Correspondingly, the actual outcome of a given supramolecular synthesis with the M-TCPP building blocks via hydrogen bonding is not always predictable.
In the above context we describe here the structures of the solid products (Fig. 1), (I) and (II), obtained by crystallizing Pd-TCPP from (i) a mixture of methanol (used to dissolve the porphyrin) and nitrobenzene (an inert reagent that commonly fills effectively intra-lattice voids in porphyrin framework solids), and (ii) a mixture of methanol and N,N'-dimethylformamide (DMF).
In the presence of alcohols (although they represent relatively weak hydrogen-bond donors and acceptors), the supramolecular hydrogen bonding of organic carboxylic acids does not always preserve the (COOH)2 intermolecular synthon. On some occasions incorporation of the hydroxy group into the intermolecular hydrogen-bonding scheme has been observed (Dale et al., 2004). Not surprisingly, the same phenomenon characterizes the hydrogen-bonded assembly in (I). Two cis-related carboxylic acid groups of Pd-TCPP are involved in R22(8) direct interaction synthons (Fig. 2a) with the carboxylic acid functions of neighboring species. However, the two other carboxylic acid residues bind to their neighbors via the R33(10) synthon (Fig. 2b), in which there is one methanol molecule inserted between the two carboxylic acid groups (Table 1). This modification maintains a continuous hydrogen-bonding pattern throughout the crystal structures, where every porphyrin unit is interacting with four different neighbouring species.
Evidently, in the present case, the hydrogen-bonded networks thus formed are no longer flat. Rather, they adopt a wavy shape, as illustrated in Fig. 3. While intermolecular aggregation via the (COOH)2 bonds is associated with coplanarity of the interacting components, that via the COOH(MeOH)COOH R33(10) synthons effects a nearly perpendicular orientation of the species involved. Thus, in the observed assemblies there is a strong kink either up or down every two rows of the interlinked Pd-TCPP units. The supramolecular networks are characterized by wide interporphyrin voids. These voids are filled in the solid phase by double interpenetration, as well as by incorporation into the structure of nitrobenzene from the solvent mixture. The crystal packing of (I) is illustrated in Fig. 4. It is further stabilized by stacking interactions at an interplanar distance of approximately 4 Å between parallel porphyrin segments of the interweaved networks.
The disruption of the interporphyrin hydrogen-bonding scheme is much more severe in (II). Here the two carboxylic acid groups of adjacent porphyrin species are solvated by two molecules of water and one molecule of DMF. One of the water molecules (O33) bridges by hydrogen bonding between two adjacent carbonyl fragments (O21 and O31), while the other water molecule (O32) and the DMF molecule (through O34) serve as H-atom acceptors from the two adjacent hydroxy residues O30 and O22 (Fig. 2c and Table 2). Moreover, every porphyrin unit connects via the first water molecule only to two neighboring porphyrins (and not to four neighbors as in the preceding example), giving rise to the formation of hydrogen-bonded one-dimensional chains as the primary supramolecular motif with coplanar porphyrin components (Fig. 5). These chain motifs further interconnect by hydrogen bonding between the O32 and O33 species of adjacent chains, yielding an extended two-dimensional hydrogen-bonded pattern in the crystal structure, which propagates parallel to the (011) plane (Fig. 5 and Table 2). The observed hydrogen-bonding scheme corresponds (when the side hydrogen bonds to the DMF molecules are ingnored) to the C33(8) graph-set representation. The packing arrangement of (II) is shown in Fig. 6. The porphyrin frameworks displaced along the a axis are arranged in an offset stacked manner with a minor overlap between adjacent molecules along the stack and average spacing of about 4.6 Å. Additional molecules of the DMF solvent are included in the interface between the hydrogen-bonded layers centered at z = 0 and z = 1 in a disordered manner (fragments O39A–C43A and O39B–C43B). They form hydrogen bonds to the O32 water molecule.
In summary, this study describes the effects of competing solvation on the self assembly of TCPP-type porphyrin building blocks by hydrogen bonding, where solvent components with comparable H-atom donating or accepting capacity interfere with direct assembly of the porphyrin units into homogeneous single-component arrays. It should be noted that Pd-TCPP and related metalloporphyrins are also excellent building blocks for networking through external metal ion bridging auxiliaries, which can coordinate to the peripheral carboxylic acid groups, and yield by multiple coordination robust supramolecular arrays (Goldberg, 2005,and references therein; Shmilovits et al., 2003; Lipstman, Muniappan, George & Goldberg, 2007).