issue contents
CCP4 Study Weekend 2023
Data - subtle details to big insights
Edited by Christoph Mueller-Dieckmann, Anna Warren and David Waterman
This virtual issue contains articles from the 2023 CCP4 Study Weekend.
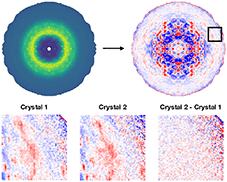
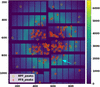
Cover illustration: Diffuse scattering is a promising method to gain additional insight into protein dynamics from macromolecular crystallography experiments. Here, diffuse patterns are visualized using the mdx2 software.
Open  access
access
 access
accessThe Guest Editors introduce the special issue based on talks at the CCP4 Study Weekend 2023. The virtual issue is available at https://journals.iucr.org/special_issues/2024/CCP42023/.
Open  access
access
 access
accessThis article introduces RGFlib, a Python package for robust statistical analysis. The package is a useful tool for a variety of tasks in X-ray crystallography data analysis, such as peak-finding, bad pixel mask making and other outlier-detection tasks.
Open  access
access
 access
accessEmerging algorithms based on machine learning offer promise in processing new diffraction experiments.
Open  access
access
 access
accessArtificial intelligence was used to characterize the diffraction in images from serial and rotation crystallography experiments. Forward simulations were used to train models to infer B factors, resolutions and the presence of crystal splitting from single diffraction images.
Open  access
access
 access
accessEMinsight is a Python-based tool for systematically mining metadata from single-particle analysis cryoEM experiments. The capture and analysis of metadata facilitates the assessment of instrument performance, provides concise reporting of experiment performance and sample quality by analysing preprocessing results, and gathers metadata for deposition. It is envisaged that this approach will benefit the microscope operator, facility managers, database developers and users.
Open  access
access
 access
accessA large set of SAD-phased PDB depositions were tested for solubility by molecular replacement using AlphaFold2 and other approaches. The 3% of cases that were not solved help to show where experimental phasing efforts could be focused in the future.
Open  access
access
 access
accessThis review covers the symptoms of radiation damage in macromolecular crystallography, how to avoid accruing radiation damage during data collection and how to identify and correct for radiation-damage artefacts in a solved structure.
Open  access
access
 access
accessmdx2 is a Python toolkit for processing diffuse scattering data from macromolecular crystals. Here, the multi-crystal scaling and merging procedures implemented in the latest version of mdx2 are described. A high-redundancy data set from cubic insulin is processed to reveal weak scattering features.
Open  access
access
 access
accessRaynals is an online, user-friendly, free (for academia) and advanced tool for the analysis of single-angle dynamic light-scattering data. Estimation of the size distribution is performed through the Tikhonov–Phillips regularization.
Open  access
access
 access
accessThe High-Pressure Freezing Laboratory for Macromolecular Crystallography (HPMX) at the ESRF allows the preparation of gas derivatives of macromolecular crystals suitable for X-ray diffraction data collection on macromolecular crystallography beamlines. Information obtained from pressurized crystals and/or gas-derivatized structures enables the improved understanding of specific issues in structural biology, such as the internal functional architecture of proteins, the interactions and reactivity of gases with macromolecules and functional structural changes including ligand-binding processes.
Open  access
access
 access
accessFrom femtoseconds to minutes: time-resolved macromolecular crystallography at XFELs and synchrotrons
This review constitutes an overview of the current status of time-resolved crystallography performed at synchrotrons and XFELs on timescales ranging from femtoseconds to minutes. Methods, potential biases, instruments and examples are presented and compared with those for the cryo-trapping of reaction-intermediate states.
Open  access
access
 access
accessThe final models for macromolecular X-ray crystallography studies are usually not only the result of refinement against some version of scaled and merged reflection data, but are often also analysed and validated purely against such merged data. Here, various examples are presented to show how the availability and use of unmerged reflection data can lead to better model analysis and improved model parametrization, as well as providing a path to better data processing and scaling.
Open  access
access
 access
accessA setup to measure time-resolved UV–Vis absorption spectra from small protein crystals using a pump–probe scheme with microsecond-to-second delays is described.
Open  access
access
 access
accessAll alternative SHELXE modes, using single or combined sources of starting phase information, are described. Side-chain tracing now completes model building in SHELXE to enhance density modification.

 journal menu
journal menu