issue contents
October 2010 issue
Joint Center for Structural Genomics (JCSG) special issue

Cover illustration: The JCSG high-throughput structural biology pipeline.
JCSG pipeline: technology and dissemination
Open  access
access
 access
accessThe Joint Center for Structural Genomics high-throughput structural biology pipeline has delivered more than 1000 structures to the community over the past ten years and has made a significant contribution to the overall goal of the NIH Protein Structure Initiative (PSI) of expanding structural coverage of the protein universe.
Open  access
access
 access
accessSpecific use cases of TOPSAN, an innovative collaborative platform for creating, sharing and distributing annotations and insights about protein structures, such as those determined by high-throughput structural genomics in the Protein Structure Initiative (PSI), are described. TOPSAN is the main annotation platform for JCSG structures and serves as a conduit for initiating collaborations with the biological community, as illustrated in this special issue of Acta Crystallographica Section F. Developed at the JCSG with the goal of opening a dialogue on the novel protein structures with the broader biological community, TOPSAN is a unique tool for fostering distributed collaborations and provides an efficient pathway to peer-reviewed publications.
domains of unknown function
Open  access
access
 access
accessDomains of unknown function (DUFs) are a large set of uncharacterized protein families that structural genomics is helping biologists to understand functionally.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessNE1406, the first structural representative of PF09410, reveals a lipocalin-like fold with features that suggest involvement in lipid metabolism. In addition, NE1406 provides potential structural templates for two other protein families (PF07143 and PF08622).
PDB reference: NE1406 from N. europaea, 2ich, r2ichsf

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of SSO2064, the first structural representative of Pfam family PF01796 (DUF35), reveals a two-domain architecture comprising an N-terminal zinc-ribbon domain and a C-terminal OB-fold domain. Analysis of the domain architecture, operon organization and bacterial orthologs combined with the structural features of SSO2064 suggests a role involving acyl-CoA binding for this family of proteins.
PDB reference: SSO2064 from S. solfataricus, 3irb

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of the first representative of DUF364 family reveals a combination of enolase N-terminal-like and C-terminal Rossmann-like folds. Analysis of the interdomain cleft combined with sequence and genome context conservation among homologs, suggests a unique catalytic site likely involved in the synthesis of a flavin or pterin derivative.
PDB reference: Dhaf4260 from D. hafniense DCB-2, 3l5o

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of the NGO1945 gene product from N. gonorrhoeae (UniProt Q5F5IO) reveals that the N-terminal domain assigned as a domain of unknown function (DUF2063) is likely to bind DNA and that the protein may be involved in transcriptional regulation.
PDB reference: NGO1945 from N. gonorrhoeae, 3dee, r3deesf

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessStructures of the first representatives of PF06684 (DUF1185) reveal a Bacillus chorismate mutase-like fold with a potential role in amino-acid synthesis.
new folds
Open  access
access
 access
accessThis review article surveys the protein structures determined by Joint Center for Structural Genomics and published in this special issue of Acta Crystallographica Section F.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of the first representative of the Pfam PF07336 (DUF1470) family reveals a two-domain organization that contains a new fold, termed the ABATE domain, at the N-terminus and a treble-clef zinc finger that is likely to bind DNA at the C-terminus.
PDB reference: Jann_2411 from Jannaschia sp. strain CCS1, 3h0n, r3h0nsf

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe first structural representative of the PF08866 (DUF1831) protein family reveals a potential new α+β fold and indicates a possible involvement in amino-acid metabolism.
PDB reference: LP2179 from L. plantarum, 2iay, r2iaysf

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessPA1994, a Pfam PF06475 (DUF1089) family homolog from P. aeruginosa, reveals remote similarities to lipoprotein localization factors and a conserved putative glycolipid-binding site.
PDB reference: PA1994 from P. aeruginosa, 2h1t, r2h1tsf

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
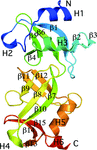
accessThe crystal structures of SPO0140 and Sbal_2486 revealed a two-domain structure that adopts a novel fold. Analysis of the interdomain cleft suggests a nucleotide-based ligand with a genome context indicating signaling as a possible role for this family.
novel variants of known folds and function
Open  access
access
 access
accessNew distinct versions of known protein folds provide a powerful means of protein-function prediction that complements sequence and genomic context analysis.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
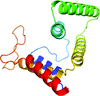
accessThe crystal structure of an essential bacterial protein, YeaZ, from T. maritima identifies an interface that potentially mediates protein–protein interaction.
PDB reference: TM0874 from T. maritima, 2a6a, r2a6asf

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of a putative NTP pyrophosphohydrolase, YP_001813558.1 from E. sibiricum, reveals a novel segment-swapped linked-dimer assembly.
PDB reference: putative NTP pyrophosphohydrolase, 3nl9

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structures of two orthologous proteins from different Shewanella species have uncovered a resemblance to CRAL-TRIO carrier proteins, which suggest that they function as transporters of small nonpolar molecules. One protein adopts an open conformation, while the other adopts a closed structure that may act as a conformational switch in the transport of ligands at the membrane surface.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessKPN03535 is a protein unique to K. pneumoniae. The crystal structure reveals that KPN03535 represents a novel variant of the OB-fold and is likely to be a DNA-binding lipoprotein.
PDB reference: KPN03535, 3f1z, r3f1zsf
human gut microbiome
Open  access
access
 access
accessMetagenomics has unleashed a deluge of sequencing data describing the organismal, genetic, and transcriptional diversity of the human microbiome. To better understand the precise functions of the myriad proteins encoded by the microbiome, including carbohydrate-active enzymes, it will be critical to combine structural studies with functional analyses.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of the BVU2987 gene product from B. vulgatus (UniProt A6L4L1) reveals that members of the new Pfam family PF11396 (domain of unknown function; DUF2874) are similar to β-lactamase inhibitor protein and YpmB.
PDB reference: BVU2987, 3due

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of BT_3984, a SusD-family protein, reveals a TPR N-terminal region providing support for a loop-rich C-terminal subdomain and suggests possible interfaces involved in sus complex formation.
PDB reference: BT_3984 from B. thetaiotaomicron, 3cgh

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of BT1062 from Bacteroides thetaiotaomicron revealed a conserved fold that is widely adopted by fimbrial components.
PDB reference: BT1062, 3gf8

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of BT2081 from B. thetaiotaomicron reveals a two-domain protein with a putative carbohydrate-binding site in the C-terminal domain.
PDB reference: putative glycoside hydrolase, 3hbz

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
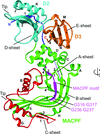
accessThe crystal structure of a novel MACPF protein, which may play a role in the adaptation of commensal bacteria to host environments in the human gut, was determined and analyzed.
PDB reference: MACPF family protein, 3kk7
ligands that aid in function characterization
Open  access
access
 access
accessAn overview and commentary on the value of liganded structures emerging from the JCSG structural genomics initiative.
Open  access
access
 access
accessA survey of the types and frequency of ligands that are bound to PSI structures is analyzed as well as their utility in functional annotation of previously uncharacterized proteins.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of the prephenate dehydrogenase component of the bifunctional H. influenzae TyrA reveals unique structural differences between bifunctional and monofunctional TyrA enzymes.
PDB reference: chorismate mutase/prephenate dehydrogenase, 2pv7

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of tryptophanyl-tRNA synthetase from T. maritima unexpectedly revealed an iron–sulfur cluster bound to the tRNA anticodon-binding region.
PDB reference: tryptophanyl-tRNA synthetase from T. maritima, 2g36

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
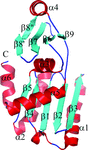
accessThe first structures from the FmdE Pfam family (PF02663) reveal that some members of this family form tightly intertwined dimers consisting of two domains (N-terminal α+β core and C-terminal zinc-finger domains), whereas others contain only the core domain. The presence of the zinc-finger domain suggests that some members of this family may perform functions associated with transcriptional regulation, protein–protein interaction, RNA binding or metal-ion sensing.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of an RmlC-type cupin with zinc acetate bound at the putative active site reveals significant differences from a previous structure without any bound ligand. The functional implications of the ligand-induced conformational changes are discussed.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessThe crystal structure of the highly specific γ-D-glutamyl-L-diamino acid endopeptidase YkfC from Bacillus cereus in complex with L-Ala-γ-D-Glu reveals the structural basis for the substrate specificity of NlpC/P60-family cysteine peptidases.
PDB reference: YkfC-l-Ala-[gamma]-d-Glu complex, 3h41
NMR in a high-throughput environment
Open  access
access
 access
accessAs an introduction to three papers on comparisons of corresponding crystal and NMR solution structures determined by the Joint Center for Structural Genomics (JCSG), an outline is provided of the JCSG strategy for combined use of the two techniques. A special commentary addresses the potentialities of the concept of `reference crystal structures', which is introduced in the following three papers.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessComparison of the NMR and crystal structures of a protein determined using largely automated methods has enabled the interpretation of local differences in the highly similar structures. These differences are found in segments of higher B values in the crystal and correlate with dynamic processes on the NMR chemical shift timescale observed in solution.
PDB reference: NP_247299.1, 2kla

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessNMR structures of the proteins TM1112 and TM1367 solved by the JCSG in solution at 298 K could be superimposed with the corresponding crystal structures at 100 K with r.m.s.d. values of <1.0 Å for the backbone heavy atoms. For both proteins the structural differences between multiple molecules in the asymmetric unit of the crystals correlated with structural variations within the bundles of conformers used to represent the NMR solution structures. A recently introduced JCSG NMR structure-determination protocol, which makes use of the software package UNIO for extensive automation, was further evaluated by comparison of the TM1112 structure obtained using these automated methods with another NMR structure that was independently solved in another PSI center, where a largely interactive approach was applied.

Loading metrics information...
Download citation


Download citation



Loading metrics information...
Open  access
access
 access
accessComparison of NMR and crystal structures highlights conformational isomerism in protein active sites
Tools for systematic comparisons of NMR and crystal structures developed by the JCSG were applied to two proteins with known functions: the T. maritima anti-σ factor antagonist TM1081 and the mouse γ-glutamylamine cyclotransferase A2LD1 (gi:13879369). In an attempt to exploit the complementarity of crystal and NMR data, the combined use of the two structure-determination techniques was explored for the initial steps in the challenge of searching proteins of unknown functions for putative active sites.


 journal menu
journal menu




















![[publBio]](http://journals.iucr.org/logos/publbio.gif)




