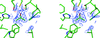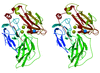issue contents
February 2001 issue
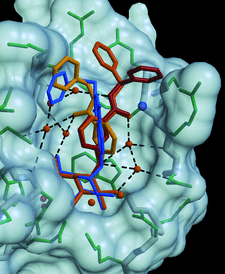
Cover illustration: Superposition of the binding modes observed for compounds BAPG (yellow, orange, red) and PEPG (blue) onto the molecular surface of the receptor-binding site of heat-labile enterotoxin, showing the range of conformations adopted in positioning the terminal phenyl ring of the ligand against the extended hydrophobic surface of the binding site in the upper portion of the figure (p. 201).
scientific comment
A survey of available databases with highly improbable strings of single amino acids is tabulated. The paper concludes with a challenge to the crystallographic community to probe the structural origins of the structure–function relationship in this neglected area.
research papers
Creatine kinase catalyses the reversible transfer of the phosphate moiety from phosphocreatine to ADP, generating creatine and ATP. The structure of a cytosolic brain-type creatine kinase at 2.3 Å is reported.
PDB reference: cytosolic bovine retinal creatine kinase, 1g0w
The X-ray structure of hydroxynitrile lyase from M. esculenta has been studied in complexes with acetone and chloroacetone. The binding observed reveals that the enzyme utilizes a general base mechanism to catalyse the reaction. In conjunction with additional kinetic data, the substrate specificity is discussed.
Heat-labile enterotoxin from E. coli (LT) and the closely related cholera toxin are responsible for a large number of severe diarrheal cases in patients worldwide. The crystallographic studies presented here show how compounds built from a galactose anchor and containing a variety of substituted phenyl-ring systems interact with the receptor-binding site of LT.
This paper presents the crystal structure determination of the RNA/DNA hybrid decamer, caaagaaaag/CTTTTCTTTG, from the polypurine tract of HIV-1 at 1.15 Å. This hybrid decamer is the largest macromolecular structure to date solved by ab initio direct methods at this lower limit of resolution.
PDB reference: RNA/DNA hybrid, 1g4q
NDB reference: AH0012
The structure of the RNA duplex r(CUGGGCGG)·r(CCGCCUGG) has been determined at 1.6 Å resolution and refined to a final R factor of 18.3% (Rfree = 24.1%).
NDB reference: r(CUGGGCGG)·r(CCGCCUGG), AR0012
The relationship between the thermostability of T. thermophilus IPMDH mutated at residue 172 and the hydrophobicity of the substituted residue is dependent on its detailed structure in the vicinity of the replaced residue.
Phasing of two proteins, the 17 kDa Fe protein myoglobin from sperm whale (P. catodon) and an 18 kDa protein (SP18) from green abalone (H. fulgens), using Kr-edge MAD with frozen crystals demonstrates the feasibility of this technique as a routine method for structure determination.
The use of short cryosoaks with salts of halides such as Br− for phasing macromolecular structures is discussed on the basis of successful experience with solving the structure of Pseudomonas carboxyl proteinase, an enzyme with molecular mass ≃ 40 kDa, by single-wavelength anomalous diffraction.
PDB reference: pepstatin-insensitive carboxyl proteinase, 1ga1
MAD phases can be complemented and improved by incorporating a direct-method procedure.
The use of dynamic light scattering to improve purification, improve crystallization and to study the self-association of full-length human replication protein A heterodimer is described.
crystallization papers
Crystals of intact tetrameric CysB from K. aerogenes have been obtained with and without its inducer N-acetylserine and in four different crystal forms.
Antimicrobial protein from P. nil has been crystallized and native data have been collected at 1.78 Å resolution using synchrotron X-rays.
Several crystal forms of the 12S central subunit of the transcarboxylase multienzyme complex from P. shermanii have been grown. Co-crystallization with either methylmalonyl CoA or malonyl CoA substrate generates crystal forms with improved diffraction and co-crystallization with CdCl2 further improves crystal stability.
The DNA binding domain of AML1 has been crystallized in space groups C2 and R32 and it was found that adding DNA improved the crystal qualities, without incorporating DNA into the crystal lattice. Crystals diffract to 1.7 and 2.0 Å in C2 and R32, respectively.
The gene for arginyl-tRNA synthetase from T. thermophilus was cloned and the recombinant protein was crystallized. X-ray data were collected from a flash-frozen native crystal to 2.3 Å resolution.
The TM1442 gene product from T. maritima has been overexpressed and crystallized (P21; a = 31.54, b = 116.83, c = 31.39 Å, β = 119.84°). Diffraction data have been collected to 2.0 Å resolution using synchrotron X-rays.
The type II dehydroquinase from H. pylori has been overexpressed and crystallized (P4232, a = b = c = 98.91 Å). Diffraction data have been collected to 2.5 Å resolution using synchrotron radiation.
CAT B2 from Tn2424, a homotrimer of 3 × 23.5 kDa, has been crystallized; the space group of the crystal is P213, with unit-cell parameter a = 130 Å. A complete data set has been collected at 2.7 Å resolution.
Crystals of γ-lactamase from C. acidovorans have been grown by the sitting-drop vapour-diffusion technique from recombinant protein expressed in E. coli. The triclinic crystals show diffraction to 2.0 Å resolution.
E. coli isopentenyl diphosphate isomerase has been produced in selenomethionyl form and the protein was crystallized by the hanging-drop vapour-diffusion method. Crystals display trigonal symmetry with unit-cell parameters a = b = 71.3, c = 61.7 Å and diffract to 1.45 Å resolution.
X-ray quality crystals have been obtained of a truncated form of cystathionine β-synthase, an enzyme involved in vascular disease which contains both a PLP and a heme cofactor.
Diffraction data to 1.6 and 2.25 Å resolution have been collected from crystals of the antisigma factor antagonist SpoIIAA from B. sphaericus in the native and phosphorylated forms, respectively.
Crystals of E. coli GlmU have been prepared in complex with coenzyme A and UDP-GlcNAc. These crystals belong to space group R32, with unit-cell parameters a = 104.5, c = 648.2 Å, and diffract X-rays to at least 2.1 Å resolution.
Outer surface protein C (OspC) from HB19 strain of B. burgdorferi has been crystallized and native data extending to 2.2 Å resolution have been collected.
A bacterial [2Fe–2S] ferredoxin possibly involved in iron–sulfur cluster biosynthesis has been crystallized for the first time. Crystals were obtained from the recombinant protein overproduced in E. coli; and a MAD experiment at the iron-edge is scheduled to solve the structure.
WPIMT, a protein methyltransferase found in wheat germ, has been purified to homogeneity and crystallized in the presence of S-adenosine-L-homocysteine. The crystals diffract to 3.3 Å and belong to the tetragonal space group P41212 or P43212.
Nine different crystal forms of dehydroquinate synthase have been obtained and characterized. Crystals of complexes with inhibitor, cofactor as well as unliganded enzyme are described.
Recombinant fructose-1,6-bisphosphate aldolase from the thermophile T. aquaticus was crystallized under two conditions. The native crystal grown under condition I diffracts to 2 Å and the SeMet crystal grown under condition II diffracts to 2.6 Å.
E. coli glucose-1-phosphatase hydrolyzes glucose-1-phosphate. The recombinant protein has been crystallized is space group R3, with two monomers in the asymmetric unit.
Crystals of the 109 C-terminal residues of the E. coli Pal gene product have been produced. Selenomethionine-substituted truncated Pal protein is currently being produced in order to solve the Pal structure using the MAD method.
Crystals of the cowpea plant phospholipase D gene product have been produced. Crystals diffract to 1.94 Å. Heavy-atom derivatives using both the calcium binding site and the phospholipid binding site of the protein are currently being produced.
Crystals of the periplasmic domain of the E. coli TolR gene product have been produced. Selenomethionine-substituted truncated Pal protein is currently produced in order to solve the TolR structure using the MAD method.
short communications
Bacterioferritin isolated from D. desulfuricans was crystallized and its structure solved using MAD data collected at the K-shell iron edge.
The structure of a 46-nucleotide RNA complex has been successfully solved using multi-wavelength anomalous dispersion at the zinc K edge.
The three-dimensional structure of the `blue' multi-copper oxidase laccase from the fungus C. cinereus at 1.68 Å reveals the structural basis for isoforms of the type 2 Cu-depleted species.
The crystal structure of an SH3 domain mutant has been refined at 1.12 Å resolution. This structure allows the interpretation on a molecular level of kinetic and thermodynamic experimental data.
PDB reference: SH3 domain mutant, 1g2b
Comparison of a new and previously determined structure of tetraubiuquitin reveals that the linkage between adjacent ubiquitin moieties is extremely flexible. This indicates that knowledge of relevant polyubiquitin conformations will require structures of polyubiquitin bound to target proteins.
PDB reference: tetraubiquitin, 1f9j
In crystallization of a Dickerson-type DNA dodecamer, a five-membered ring of water molecules can replace the hydrated magnesium cation to link neighboring duplexes in the crystal lattice, although the magnesium cation is more effective and gives X-ray diffraction at slightly higher resolution.
meeting reports
Free 

The current state of the art and future perspectives for protein crystallography with neutrons are reviewed and the relevance of neutrons in biology discussed briefly.


 journal menu
journal menu