issue contents
August 2003 issue

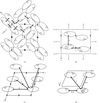
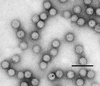
Cover illustration: The packing diagram for histone octamers in crystals having space group P65, viewed down the c axis. The packing results from interactions between the interpenetrating left-hand-helical arrays of the octamers (p. 1395).
research papers
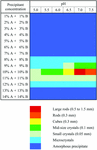
The strengths of intermolecular contacts were calculated for polymorphous lysozyme crystals. The surface energies were calculated to correlate with morphology.
The crystal structure of cod uracil-DNA glycosylase determined at 1.9 Å resolution reveals a decreased number of ion-pair interactions, a less optimally packed core and an optimization of the electrostatic surface potential of the DNA-binding regions compared with the human counterpart.
PDB reference: uracil-DNA glycosylase, 1okb, r1okbsf
The crystal structure of β-luffin at 2.0 Å resolution has been solved by the molecular-replacement method using polyalanyl trichosanthin as the search model.
PDB reference: β-luffin, 1nio, r1niosf
The three-dimensional `fast rotational matching' method is extended to the full six-dimensional (rotation and translation) matching scenario between two three-dimensional objects. The approach allows treatment of five of the six degrees of freedom by means of fast Fourier transforms.
The crystal structure of a DNA/RNA chimera (dC)(rC)d(ATCG) complexed with daunomycin adopts the B-form helical conformation, similar to the DNA counterpart duplex. A model of B-form RNA duplex has been suggested, in which the interaction of the 2′-hydroxyl groups depends on their 3′-side nucleotide.
PDB reference: (dC)(rG)d(ATCG)–daunomycin complex, 1jo2, r1jo2sf
NDB reference: DD0041
In the absence of an ordered water structure in a crystal air-dried at room temperature, the N-termini of the A chains undergo partial unfolding. Both at room temperature and at 100 K, the majority of the GluB13 side chains in the central core of the hexamer in the air-dried crystal are disordered.
The structure of the histone-core octamer crystallized in KCl/phosphate compares well with the octamer in the nucleosome but exhibits unique inter-octamer crystal-packing interactions.
PDB reference: histone-core octamer, 1hq3, r1hq3sf
A method for manual crystallization using nanolitre volumes is reported.
The structure of the LanD protein MrsD at 2.54 Å is described.
PDB reference: MrsD, 1p3y, r1p3ysf
The light-harvesting I complex was found to be most soluble in detergent and amphiphile combinations that also yielded protein crystals. Amphiphiles that were effective for crystallization were found by mass spectrometry to displace bound detergent.
Low-redundancy SAD data recorded at λ = 2.0 Å on a triiodide derivative of porcine pancreatic elastase shows evidence of radiation damage at iodine heavy-atom sites which can be used very simply within SHARP to enhance the phasing signal.
The structures of two crystal forms of a four-helix bundle protein have been solved using group–subgroup relations between space groups of polymorphic protein crystals.
A practical procedure for MAD phasing was successfully performed without the aid of XAFS measurements to solve the crystal structure of the outer-membrane lipoprotein carrier LolA from E. coli.
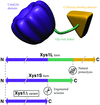
The three-dimensional structure of the catalytic domain is reported at 1.78 Å resolution, showing a (β/α)8 barrel fold, and compared with related xylanases.
PDB reference: Xys1Δ, 1nq6, r1nq6sf
The crystal structure of trimeric KDPG aldolase from P. putida, a Class I Schiff-base forming enzyme in the Entner–Doudoroff pathway, is described.
PDB reference: KDPG aldolase, 1mxs, r1mxssf
crystallization papers
A four-copper-containing laccase from C. hirsutus was crystallized. The data were collected to 1.85 Å resolution and initial phases have been obtained using the SAD method.
The extrinsic protein in an oxygen-evolving complex (OEC23) of photosystem II, tobacco OEC23, was crystallized by the hanging-drop vapour-diffusion technique with PEG 4000 and diffraction data were collected to 2.0 Å.
LC3, a protein involved in autophagy in mammalian cells, was crystallized. A complete diffraction data set was collected to a resolution limit of 2.1 Å.
Crystals of the C-terminal truncated domain of the S-layer protein SbsC from G. stearothermophilus have been obtained. Diffraction data were collected to a resolution of 3.0 Å from a native crystal and to 3.2 Å from a Pt derivative.
Glutathione S-transferase from the human malarial parasite P. falciparum has been crystallized, allowing its X-ray structure determination.
The carboxylesterase Est30 from B. stearothermophilus has been crystallized in space group C2221, with unit-cell parameters a = 55.83, b = 58.15, c = 179.65 Å. The crystal diffracted to better than 2.0 Å resolution.
Recombinant protein mavicyanin from C. pepo medullosa, which is a member of the phytocyanin subclass of cupredoxins was crystallizaed and its preliminary crystallographic data were reported.
Human uridine-cytidine kinase 2 alone or in complex with ligands crystallized in six crystal forms. One form diffracted to 1.8 Å resolution.
An autoinhibited form of the tandem SH3 domain of p47phox, phagocyte NADPH oxidase regulatory component, has been crystallized and X-ray diffraction data have been collected to 2.1 Å resolution using synchrotron radiation.
The hibiscus chlorotic ringspot virus (HCRSV), a member of the family Tombusviridae, has been purified and subsequently crystallized in space group P23.
An intramolecular complex consisting of the interacting domains from two transcriptional regulators, LMO4 and ldb1, has been crystallized. Data were collected to 1.3 Å resolution in addition to MAD data to 1.7 Å resolution.
Crystals of the fungal immunomodulatory protein Fve from the edible golden needle mushroom (F. velutipes) have been obtained and diffracted to 1.4 Å resolution.
KsgA, a ubiquitous rRNA dimethyltransferase, has been crystallized in space group C2, with unit-cell parameters a = 173.9, b = 38.4, c = 83.0 Å, β = 90.0°. X-ray data were collected to 1.9 Å with Cu Kα radiation.
The crystallization of the tetrameric type IIF restriction endonuclease SfiI in complex with DNA is reported.
Human pirin, a novel highly conserved nuclear protein, has been cloned, expressed, purified and crystallized using PEG as precipitant.
The crystallization and preliminary X-ray characterization of alginate lyase (family PL-7) from P. aeruginosa are presented.
Human DJ-1, a protein associated with male fertility and parkinsonism has been crystallized. A data set was collected to 2.00 Å resolution.
The structural domain of the PRRSV nucleocapsid N protein has been expressed in E. coli and crystallized and a complete native data set has been collected at 2.7 Å resolution.
A putative dTDP sugar epimerase (NovW) from S. spheroides, implicated in novobiocin biosynthesis, has been overexpressed in E. coli and purified and crystals diffracting X-rays to 2.0 Å resolution have been obtained.
MurC is responsible for the first step in the addition of the pentapeptide unit of peptidoglycan. Native and SeMet crystals of the E. coli enzyme have been prepared.
The 36 kDa relaxase domain of the TraI protein of conjugative plasmid F factor has been crystallized by the sitting-drop method in native and selenomethionine-substituted forms. Data sets from trigonal crystals have been collected to 2.5 Å resolution for the native protein and to 3.1 Å resolution for the selenomethionine protein.
Phosphotransacetylase has been crystallized for the first time. The enzyme from M. thermophila was crystallized in space group I41, with unit-cell parameters a = b = 114.8, c = 127.8 Å, α = β = γ = 90°; the crystals diffracted to 2.5 Å resolution.
short communications
The C-terminal helix involved in RNA binding has been shown to occlude the binding surface in a previous solution structure of a free U1A. A new 1.8 Å resolution structure of the free protein shows the helix in the same location as in an RNA-bound complex.
PDB reference: U1A RNA-binding domain, 1nu4, r1nu4sf
5-Iodo-2′-deoxyuridine (IDU) was the first antiviral nucleoside against herpes simplex virus type 1 and type 2. The structure of d[CACG(IDU)G] reveals two cobalt hexammine complexes in binding modes similar to other cobalt hexammine-containing Z-DNA fragments.
PDB reference: d[CACG(IDU)G], 1omk, r1omksf
Conformational change and hydrogen bonding of protein-bound astaxanthin are argued to be the predominant factors responsible for the purple–blue colour of the lobster carapace carotenoprotein β-crustacyanin. The further shifted visible spectra of carotenoids with a cyclopentenedione ring remain to be accounted for.
books received
Free 

Free 



 journal menu
journal menu