issue contents
April 2007 issue
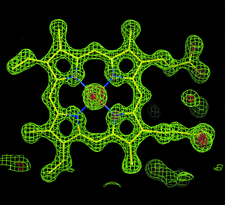
Cover illustration: 2Fo - Fc map for the CN- bound form of Arthromyces ramosus peroxidase at 1.3 Å resolution (contour level: green 2![[sigma]](/logos/entities/sigma_rmgif.gif) , red 6
, red 6![[sigma]](/logos/entities/sigma_rmgif.gif) )(p. 472).
)(p. 472).
research papers
The crystal structure of L. lactis CcpA is reported.
PDB reference: CcpA, 2o20, r2o20sf
The crystal structures of four complexes of a signalling glycoprotein from goat (SPG-40) with oligosaccharides of different lengths have been determined. The structures of the complexes with GlcNAc3, GlcNAc4, GlcNAc5 and GlcNAc6 revealed the positions of these oligosaccharides in the carbohydrate-binding groove and provided insights into the binding mechanism of oligosaccharides to SPG-40.
An automation pipeline for macromolecular structure solution by molecular replacement with a special emphasis on the discovery and preparation of a large number of search models is described. It is concluded that exploring a range of search models automatically can be valuable in many cases.
Packing and loop-structure variations in non-isomorphous crystals of FabZ from Plasmodium falciparum
The crystal structures of FabZ from P. falciparum in two crystal forms are reported and a detailed structural comparison is made of three crystal forms of this enzyme in order to explore the possible reasons for the existence of non-isomorphism.
The crystal structures of the separate dimerization domains of the periplasmic disulfide-bond isomerases DsbC and DsbG have been determined at resolutions of 2.0 and 1.9 Å, respectively.
Crystal structures of CN−-, NO- and hydroxylamine-bound forms of A. ramosus peroxidase at 1.3 Å resolution show the coordination geometries of the ligands to the haem iron.
The structure of the protein-targeting chaperone CsaA from B. subtilis has been solved to 2.0 Å resolution from a trigonal crystal form (P3221) and to 1.9 Å resolution from a tetragonal crystal form (P4212).
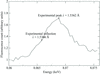
Comparison of diffraction data collected from matched protein crystals bathed in helium at a set temperature of 8 K and nitrogen at 100 K indicate that the use of cryogenic helium can increase the diffractive lifetime of crystals and measurably mitigate radiation damage.
The application of ADP-2Ho as a heavy-atom derivative for the structure determination of MTR kinase is described.
PDB reference: MTR kinase–ADP-2Ho complex, 2olc, r2olcsf
The crystal structure of trypsin with a designed highly potent synthetic inhibitor has been determined at atomic resolution. The structure has revealed the precise nature of the interactions between the enzyme and the inhibitor molecules.
PDB reference: trypsin–inhibitor complex, 2ayw, r2aywsf
Crystal structures were determined of the extracellular part of glutamate carboxypeptidase II (GCPII; amino-acid residues 44–750) in complex with two potent inhibitors, quisqualate and 2-PMPA (the most potent GCPII inhibitor to date), at resolutions of 3.0 and 2.2 Å, respectively.
The program CAALIGN performs pairwise structure alignment and multiple structure alignment of protein structures based on the maximal superposition of Cα coordinates.
A method is described whereby the addition of a nonspecific fluorescent dye to a wide variety of different protein solutions prior to crystallization results in a significantly increased fluorescence signal being obtained from protein crystals. Subsequent fluorescence imaging of the crystallization experiments demonstrates that successful experiments containing crystals may be easily discriminated from other objects produced by the crystallization experiment.
Crystal structures and cross-linking experiments of the N- and C-terminal domains of T. maritima trigger factor (TF) are presented. Structural and cross-linking data suggest that TF could interact with substrate proteins via hydrophilic surfaces.
short communications
The noncrystallographic symmetry operations reported for two crystal structures of phospholipase A2 isoforms from the common krait can be reinterpreted as crystallographic operations. The change in space group affects conclusions about the oligomeric forms of the proteins in the crystalline state.
PDB reference: phospholipase A2, 2osn, r2osnsf
The formation of nuclei in a crystallization experiment requires the interaction of protein molecules until a critical size of aggregate is created. The formation of such nuclei may be achieved either at very high protein concentrations or at lower levels of saturation with the introduction of seeds to form nucleation sites. Incorporating this seeding step into an automated screening process could have a considerable influence on the number of conditions which produce crystals. Some preliminary results are presented in this article.


 journal menu
journal menu