issue contents
August 2019 issue
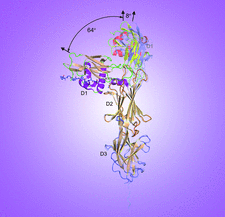
Cover illustration: Structures of major pilins in Clostridium perfringens demonstrate dynamic conformational change [Tamai et al. (2019), Acta Cryst. D75, 718-732]. Pili in Gram-positive bacteria are flexible rod proteins associated with the bacterial cell surface which play important roles in the initial adhesion to host tissues and colonization. The pilus shaft is formed by the covalent polymerization of major pilins, catalyzed by sortases, a family of cysteine transpeptidases. X-ray structures of the major pilins from C. perfringens strains 13 and SM101, and of sortase from strain SM101 are published with biochemical analysis to detect the formation of pili in vivo.
research papers
Open  access
access
 access
accessPeakProbe facilitates the automated modelling of ordered solvent in macromolecular crystal structures by analysing features of the electron density and chemical environment surrounding a given coordinate. The extracted data are transformed to a resolution-independent score space and likely solvent models are predicted based on the frequency distributions observed in a large-scale sample of the PDB.
Pili, polymers of the major pilin, in Gram-positive bacteria are flexible rod proteins associated with the bacterial cell surface that play important roles in initial adhesion to host tissues and colonization. Crystal structures of major pilins from Clostridium perfringens strains have been determined, which were considered to be in an equilibrium state between an elongated and a bent structure through dynamic conformational change.
Open  access
access
 access
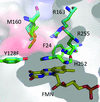
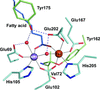
accessStructural and enzymological explorations of α-hydroxyacid oxidases uncover new flavin mononucleotide-mediated reactions and intermediates.
PDB references: p-hydroxymandelate oxidase, 5zzp; complex with 4-hydroxy-(S)-mandelate, 5zzq; complex with (S)-mandelate, 5zzr; complex with benzoate, 5zzs; complex with 2-phenylpropanoate, 6a00; complex with benzoylformate, 6a08; Y128F mutant, 6a13; complex with (R)-mandelate, 5zzx; complex with 2-hydroxypropanoate, 5zzy; complex with 2-phenylpropanoate, 6a0d; complex with 2-hydroxy-3-phenylpropanoate, 6a0g; complex with 2-phenylacetate, 6a0m; complex with benzaldehyde, 6a0o; complex with (S)-mandelate, 6a0v; complex with benzoate, 6a0y; complex with phenylpyruvic acid, 6a11; complex with benzoylformate, 6a19; complex with 4-hydroxy-(S)-mandelate, 6a1a
A new haloacid dehalogenase (HAD) phosphatase originating from Thermococcus thioreducens was expressed, purified and crystallized, and its structure was refined to 1.75 Å resolution. Structural studies confirmed the presence of the canonical HAD phosphatase fold with the HAD signature motifs and of Mg2+ ion in the active site. The results of computational docking and enzymatic assays identified possible substrates of the enzyme.
PDB reference: phosphatase Tt82 from Thermococcus thioreducens, 6iah
Open  access
access
 access
accessRecent advances in automated protein model building using ARP/wARP are presented. The new methods include machine-learning-enhanced sequence assignment and loop building using a fragment database.
A method is described for the quantification of the relative amounts of different metal ions in the binding sites of metalloproteins using X-ray anomalous dispersion.
The CAMP factor of group B streptococcus, a pore-forming toxin, was crystallized with an MBP tag. Structural and functional analysis suggests that the basic patch formed by helices 6 and 7 within the C-terminal domain is critical for its activity.
PDB reference: MBP-CAMP, 5y2g


 journal menu
journal menu