issue contents
November 2022 issue

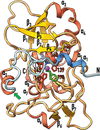
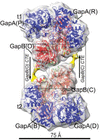
Cover illustration: Photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a key enzyme in the Calvin–Benson cycle [Marotta et al. (2022), Acta Cryst. D78, 1399–1411]. A GAPDH oligomer cryo-EM density map fitted with a model derived from the crystal structure of oxidized A2B2 complexed with NADP+ is shown. This 13 Å resolution A10B10 map is a pentamer of A2B2 tetramers with C5 symmetry and a central 5531 Å2 wide seastar-shaped cavity.
CCP4
Open  access
access
 access
accessThe accuracy of the models predicted using AlphaFold and RoseTTAFold eases phasing with molecular replacement and raises the question of model bias. ARCIMBOLDO_SHREDDER solves structures while aiming to establish the experimental information in a crystallographic determination by introducing systematic, local verification of phasing solutions.
Open  access
access
 access
accessA brief overview of techniques used in the hit-identification/validation stage of drug discovery is given, highlighting the importance of orthogonal measurements and triangulation in early-stage crystallographic hit validation.
Open  access
access
 access
accessphenix.process_predicted_model and ISOLDE provide seamless integration of AlphaFold-predicted models into protein structure-solution workflows.
research papers
Open  access
access
 access
accessSmall-angle X-ray scattering (SAXS) and small-angle neutron scattering (SANS) measurements of five standard proteins in solution using 12 SAXS and four SANS instruments demonstrate reproducibility and yield consensus scattering profiles that provide a foundation benchmarking set to evaluate approaches to scattering-profile prediction from atomic coordinates.
SASBDB references: SAXS data: ribonuclease A, SASDPP4; urate oxidase, SASDPQ4; xylose isomerase, SASDPR4; xylanase, SASDPS4; lysozyme, SASDPT4; SANS data: ribonuclease A in D2O buffer, SASDPU4; lysozyme in D2O buffer, SASDPV4; xylanase in D2O buffer, SASDPW4; urate oxidase in D2O buffer, SASDPX4; xylose isomerase in D2O buffer, SASDPY4; lysozyme in H2O buffer, SASDPZ4; ribonuclease in H2O buffer, SASDP25; xylanase in H2O buffer, SASDP35; urate oxidase in H2O buffer, SASDP45; xylose isomerase in H2O buffer, SASDP55
Open  access
access
 access
accessCoating cryo-EM grids with single-stranded DNA before applying the sample reduces the aggregation of macromolecules and improves their distribution. It enables the determination of structures from samples that aggregate on conventional grids.
Open  access
access
 access
accessThe horseshoe crab Limulus polyphemus is an ancient chelicerate that is a model organism for the study of the evolution and function of peptidases. It contains a member of the astacin metallopeptidases, the mechanism of latency of which was revealed by X-ray structural analysis.
PDB reference: Limulus polyphemus astacin zymogen, 8a28
A phylogenetically distant glycoside hydrolase family 5 member retrieved from the capybara gut microbiome displays a large and open catalytic interface that is adapted to recognize complex heteromannans, establishing the novel GH5_57 subfamily. This study expands the current mechanistic and functional understanding of one of the largest GH families.
PDB reference: CapGH5_57, 8d89
Open  access
access
 access
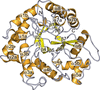
accessThe crystal structure of human AAR2 bound to the central spliceosomal factor PRPF8 and in vitro functional data yield insights into the structural basis of snRNP assembly in humans.
PDB reference: AAR2 bound to PRPF8 RNaseH domain, 7pjh
Open  access
access
 access
accessThe crystal structure of DNA polymerase I from Thermus phage G20c is presented and represents the first crystal structure of DNA polymerase I from a group of related thermophilic bacteriophages. The structure reveals a new structural motif termed SβαR that was not previously identified in other DNA polymerases I (or in the DNA polymerase A family).
Regulation of the heteromeric form of photosynthetic glyceraldehyde 3-phosphate dehydrogenase (AB-GAPDH) depends on oscillation between a fully active heterotetramer (A2B2) and inhibited oligomers. Experimental evidence demonstrates that the inhibition of spinach AB-GAPDH depends on the formation of dimers, tetramers or pentamers of A2B2 modules linked together by C-terminal extensions of the B subunits that extend from one modular tetramer and occupy two active sites of the adjacent tetramer.


 journal menu
journal menu