issue contents
October 2022 issue

Cover illustration: The quorum-sensing regulator, RdfS, forms a helical filament with eightfold screw symmetry [Verdonk et al. (2022), Acta Cryst. D78, 1210–1220].
CCP4
Open  access
access
 access
accessAn introduction to the Proceedings of the 2020 CCP4 Study Weekend on model building which are available as a virtual issue at https://journals.iucr.org/special_issues/2020/CCP42020/.
research papers
Differences between the binding of RNA and DNA in S1 nuclease were analysed using X-ray crystallography. In order to properly interpret the obtained diffraction data, a correction for multiple lattice-translocation defects was applied.
Open  access
access
 access
accessThe X-ray crystallographic structure of RdfS reveals molecular superhelical polymers in the crystal.
PDB reference: RdfS, 8dgl
SASBDB reference: RdfS, SASDPK4
Open  access
access
 access
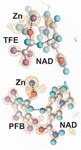
accessThe crystallographic temperature factors determined for three different complexes of horse liver alcohol dehydrogenase over the range 25–150 K are relatively independent of temperature and suggest that the B factors at 100 K are relevant to catalysis at 300 K.
PDB references: alcohol dehydrogenase, complex with NAD and TFE, 25 K, 7ua6; 45 K, 7uc9; 65 K, 7uca; 85 K, 7ucu; 100 K, 4dxh; 125 K, 7ude; 150 K, 7udd; complex with NAD and PFB, 25 K, 7udr; 50 K, 7uec; 75 K, 7uee; 85 K, 7uef; 100 K, 7uei; 150 K, 7uej; G173A alcohol dehydrogenase, complex with NAD and PFB, 50 K, 7uhv; 85 K, 5kj1; 120 K, 7uhw; 150 K, 7uhx
The structure of the elaiophylin glycosyltransferase homodimer reveals a novel model of a self-activating form of a member of glycosyltransferase family 1 that binds the symmetric substrate.
Open  access
access
 access
accessA computational method for obtaining the time-dependent intermediate scattering function of supported membrane stacks is presented.
Open  access
access
 access
accessThis study describes the structure of the chloroplast ribosome-associated molecular chaperone trigger factor at 2.6 Å resolution. It is shown that this eukaryotic trigger factor has evolved specific structural features in plants that are distinct from those of the bacterial homolog, contributing to a better understanding of co-translational protein folding in plastids.
PDB reference: chloroplast trigger factor, 7zgi
The structural basis for the reduction of berberine by bacterial nitroreductases is described.
PDB reference: Escherichia coli NfsB in complex with berberine, 7x32


 journal menu
journal menu