issue contents
December 2023 issue
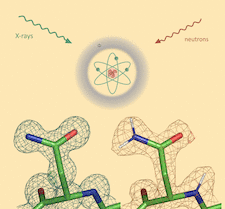
Cover illustration: Appearance of an ASN residue in electron (left) and nuclear scattering length (right) density maps [Liebschner et al. (2023), Acta Cryst. D79, 1079–1093]. The orientation of the side chain is often ambiguous in electron density maps. However, if the H atoms of the head group are replaced by D, the NH2 (ND2) group turns into a stronger scatterer than the O atom in nuclear scattering length density maps. This makes the orientation of the side-chain much more discernible.
CCP4
Open  access
access
 access
accessThe macromolecular refinement package REFMAC5 from the CCP4 suite has been extended by the incorporation of algorithms for neutron crystallography.
Open  access
access
 access
accessThe explicit refinement of Ramachandran, rotamer and clash criteria at now-prevalent lower resolutions (2.5–4 Å) has made the current, traditional model validation at the Protein Data Bank nearly useless in this range, since quite poor structures can have perfect scores. Fortunately, new criteria and programs such as ISOLDE, CaBLAM and AlphaFold are coming to the rescue, are already very useful and should be extensible into an effective new community standard.
Open  access
access
 access
accessThe improved joint X-ray and neutron refinement procedure in Phenix optimizes two different models against the X-ray and neutron data sets. This approach is shown to reduce overfitting compared with refining the models separately.
research papers
The crystal structures of different states of cyanase, i.e. native, cyanate-bound, bicarbonate-bound and reaction states, obtained using synchrotron X-rays and X-ray free-electron lasers, elucidate the reaction intermediates and molecular dynamics of the active site.
Open  access
access
 access
accessThe components of the graphlet degree vector, which describes the complexity of the wiring of a given atom, can be used in a multiple linear regression model to predict atomic displacement parameters in protein structures.

 journal menu
journal menu