issue contents
September 2014 issue
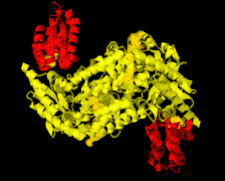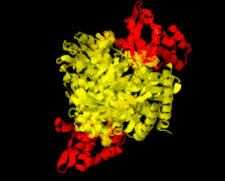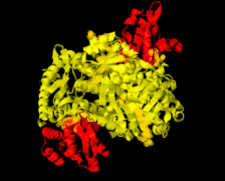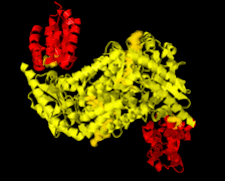
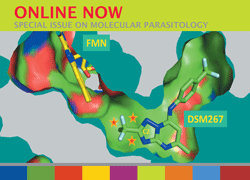
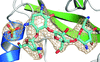
Cover illustration: Nfs2-SufE1 complex involved in sulfur mobilization for iron-sulfur cluster biogenesis in Arabidopsis thaliana plastids (Roret et al., p. 1180).
IYCr crystallization series
Microseed matrix screening is increasingly successful in the optimization of protein crystallization leads.
structural communications
Open  access
access
 access
accessCrystals of HEWL with cisplatin and HEWL with carboplatin grown in sodium iodide conditions both show a partial chemical transformation of cisplatin or carboplatin to a transiodoplatin (PtI2X2) form. The binding is only at the Nδ atom of His15. A further Pt species (PtI3X) is also seen, in both cases bound in a crevice between symmetry-related protein molecules.
Open  access
access
 access
accessThe platinum hexahalides have an octahedral arrangement of six halogen atoms bound to a Pt centre, thus having an octahedral shape that could prove to be useful in interpreting poor electron-density maps. In a detailed characterization, PtI6 chemically transformed to a square-planar PtI3 complex bound to the Nδ atom of His15 of HEWL was also observed, which was not observed for PtBr6 or PtCl6.
Open  access
access
 access
accessAn X-ray crystal structure showing the binding of purely carboplatin to histidine in a model protein has finally been obtained. This required extensive crystallization trials and various novel crystal structure analyses.
PDB references: HEWL co-crystallized with carboplatin in MPD conditions: crystal 1 processed using the EVAL software package, 4lt0; carboplatin binding to HEWL in NaBr crystallization conditions studied at an X-ray wavelength of 0.9163 Å, 4nsf; HEWL co-crystallized with carboplatin in MPD conditions: crystal 2 processed using the XDS software package, 4lt3; carboplatin binding to HEWL in 20% propanol, 20% PEG 4000 at pH 5.6, 4nsi; carboplatin binding to HEWL in NaBr crystallization conditions studied at an X-ray wavelength of 1.5418 Å, 4nsg; carboplatin binding to HEWL in 0.2 M ammonium sulfate, 0.1 M sodium acetate in 25% PEG 4000 at pH 4.6, 4nsh; carboplatin binding to HEWL in 2 M ammonium formate, 0.1 M HEPES at pH 7.5, 4nsj
Open  access
access
 access
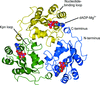
accessBMN 673, a novel PARP1/2 inhibitor in clinical development with substantial tumor cytotoxicity, forms extensive hydrogen-bonding and π-stacking in the nicotinamide pocket, with its unique disubstituted scaffold extending towards the less conserved edges of the pocket. These interactions might provide structural insight into the ability of BMN 673 to both inhibit catalysis and affect DNA-binding activity.
The crystal structure of shrimp nucleoside diphosphate kinase in binary complex with dCDP, dADP and ADP shows conserved contacts independent of the kind of nucleotide bound.
Two alternate crystal forms of the human CD44 hyaluronan-binding domain, one hexagonal and one monoclinic, have been determined at 1.60 and 1.08 Å resolution, respectively. A short peptide found in the monoclinic form may identify a site of CD44 for protein–protein interactions.
The structure of Na-GST-1 in complex with GST is reported.
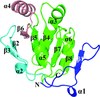
In this study, the crystal structure of a DsbE or DsbF homologue protein from Corynebacterium diphtheriae has been determined, which revealed a thioredoxin-like domain with a typical CXXC active site.
PDB reference: DsbF homologue, 4pq1
The mycobacterial Eis proteins are aminoglycoside acetyltransferases. Here, the crystal structure of M. smegmatis Eis in the paromomycin-bound form is reported at 3.3 Å resolution.
PDB reference: Eis, complex with paromomycin, 4qb9
Cysteine desulfurases (CDs) are enzymes that provide sulfur for several biological processes, including the synthesis of iron–sulfur clusters. The difference in the capacity of the two existing groups of CDs to interact with various partners seems to depend on the flexibility of an extended lobe that harbours the catalytic cysteine.
Open  access
access
 access
accessThe structure of T. gondii fructose-1,6-bisphosphate aldolase, a glycolytic enzyme and structural component of the invasion machinery, was determined to a resolution of 2.0 Å.
PDB reference: fructose-1,6-bisphosphate aldolase, 4tu1
The structure of Xsa43E possesses the characteristic five-bladed β-propeller domain seen in all GH43 enzymes. The functional characterization of Xsa43E shows it to be an arabinofuranosidase capable of cleaving arabinose side chains from short segments of xylan.
PDB reference: Xsa43E, 4nov
Open  access
access
 access
accessThe crystal structure of A. baumannii alanine racemase (AlrAba) from the highly antibiotic resistant NCTC13302 strain has been solved to 1.9 Å resolution. Comparison of (AlrAba) with alanine racemases from closely related bacteria demonstrates a conserved overall fold.
PDB reference: alanine racemase, 4tlo
crystallization communications
The expression, purification, crystallization, preliminary X-ray crystallographic and cryo-EM analysis of the bifunctional enzyme fucokinase/L-fucose-1-P-guanylyltransferase are reported. A diffraction data set was collected to 3.7 Å resolution from the full-length native protein crystal using synchrotron X-rays. The preliminary cryo-EM image showed two parallel disc-shaped objects with a diameter of 10 nm.
The redox domain of APR1 from A. thaliana was crystallized in the tetragonal space group P43212 or P41212 and diffraction data were collected to 2.70 Å resolution.
The periplasmic domain of FliP of the bacterial flagellar type III export apparatus has been expressed, purified and crystallized, and the crystals have been characterized by X-ray diffraction.
Open  access
access
 access
accessThe periplasmic domain of the E. coli aspartate receptor Tar was cloned, expressed, purified and crystallized with and without bound ligand. The crystals obtained diffracted to resolutions of 1.58 and 1.95 Å, respectively.
The cloning, overexpression, purification, crystallization and preliminary crystallographic analysis of the C-terminal domain of prostate apoptosis response-4 protein (Par-4) are presented.
The purification, crystallization and preliminary X-ray crystallographic analysis of B. cereus glycosyltransferase-1 are reported.
Aiming at the identification of new carbohydrate-binding modules (CBMs), a sugarcane soil metagenomic library was analyzed and an uncharacterized CBM (CBM_E1) was identified. In this study, CBM_E1 was expressed, purified and crystallized and X-ray diffraction data were collected to 1.95 Å resolution.
Open  access
access
 access
accessE. coli YfcM was expressed, purified and crystallized. Crystals of YfcM were obtained by the in situ proteolysis crystallization method. Using these crystals, an X-ray diffraction data set was collected at 1.45 Å resolution.
Human myotubularin-related protein 3 was cloned, expressed, purified and crystallized. X-ray diffraction data were collected to 3.3 Å resolution.
Monodehydroascorbate reductase (OsMDHAR) from O. sativa L. japonica was expressed as the active form using E. coli strain NiCo21 (DE3). X-ray diffraction data were collected to 1.9 Å resolution.
The glucose-1-phosphate uridylyltransferase from E. amylovora has been cloned, expressed, purified and crystallized. Data were collected to 2.46 Å resolution in space group P62 and the structure was solved by molecular replacement using the structure of the enzyme from E. coli.
C-terminal domains 2 and 3 of Ski7 from S. cerevisiae were purified and crystallized. The crystals diffracted to 2.0 Å resolution.
Preliminary crystallographic analysis of the 16S rRNA methyltransferase RsmI from E. coli is reported.
The crystallization of TssL from V. cholerae is reported.
A crystal of the chicken MHC class I molecule BF2*0401 in complex with chicken CD8αα (CD8αα–BF2*0401) diffracted to 2.8 Å resolution and belonged to space group P21, with unit-cell parameters a = 90.6, b = 90.8, c = 94.9 Å, β = 98°.
The expression, purification and crystallization of Xaa-Pro dipeptidase from X. campestris (GenBank accession No. NP_637763) and its preliminary X-ray diffraction analysis to 1.83 Å resolution are reported.
The C-terminal DUF490963–1138 domain of TamB (previously YtfN) from E. coli strain K-12 has been overexpressed, purifed and crystallized. Preliminary diffraction data have been collected to 2.1 Å resolution.
The N-acetyltransferase PseH from the H. pylori pseudaminic acid biosynthesis pathway has been purified and co-crystallized with acetyl-CoA by the hanging-drop vapour-diffusion method. A diffraction data set has been collected to 2.5 Å resolution.
Crystallization and preliminary X-ray crystallographic analysis of the PH-GRAM domain of human MTMR4
The PH-GRAM domain of human MTMR4 was cloned, expressed, purified and crystallized. X-ray diffraction data were collected to 3.20 Å resolution.
A new crystal form of the complex of E. coli O157 PaaA2 and ParE2 is described. The crystals belong to space group P41212 (or P43212) and diffract to 2.8 Å resolution. SeMet-derivatized crystals of the complex diffracting to the same resolution were successfully obtained. Finally, crystallization of the complex of a PaaA2 truncation and ParE2 is also described.
The expression, purification and crystallization of acylpeptide hydrolase from D. radiodurans (GenBank accession No. NP_293889) and its preliminary X-ray diffraction analysis to 2.6 Å resolution are reported.
laboratory communications
A survey of the Protein Data Bank for entries with a large number of molecules in the asymmetric unit was performed and the strategies used for molecular replacement are described.


 journal menu
journal menu














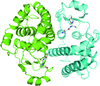

















![[publBio]](/logos/publbio.gif)