issue contents
January 2017 issue
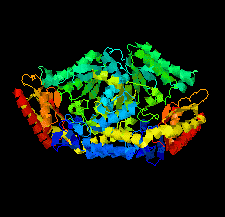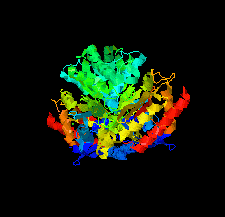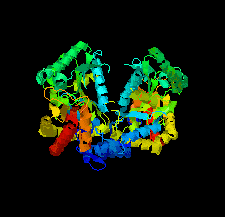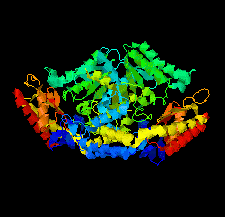
Cover illustration: Crystal structure of an industrially relevant enzyme, putrescine aminotransferase from Pseudomonas sp. strain AAC (Wilding et al., p. 29).
research communications
Selective targeting of the ζ1 subunit of the human coatomer protein complex I (Copζ1) is a promising avenue for anticancer therapy, as knockouts of ζ1 have been shown to specifically kill both proliferating and nondividing tumour-cell populations. In order to provide a structural basis for rational drug design, the high-resolution crystal structure of Copζ1 is reported.
PDB reference: Copζ1, 5mc7
The C-terminal domain (residues 780–1122) of the ATPase motor Fun30 from S. cerevisiae has been crystallized and the structure was determined at a resolution of 1.95 Å. A helix-bundle insertion in the ATPase domain was identified to be specific to the Fun30 subfamily.
PDB reference: C-terminal part of Fun30 ATPase domain, 5gn1
The crystal structure of an IclR homologue from Microbacterium sp. strain HM58-2 was determined at 2.1 Å resolution. It revealed a unique tetramer conformation, which may represent one of the multiple conformations in transcriptional regulation.
PDB reference: Mi-IclR, 5h1a
The crystal structure of a malonate-semialdehyde dehydrogenase from Pseudomonas sp. strain AAC has been determined to a nominal resolution of 2.95 Å.
PDB reference: malonate semialdehyde dehydrogenase, 5tjr
The crystal structure of a putrescine aminotransferase from Pseudomonas sp. strain AAC has been determined to a resolution of 2.07 Å.
PDB reference: putrescine aminotransferase, 5ti8
The structure of a new fungal form of aspartate-semialdehyde dehydrogenase from A. fumigatus reveals differences in secondary-structural features that will be exploited for species-selective antibiotic development.
PDB reference: A. fumigatus ASADH, 5jw6
The extracellular domain of human hepatocyte growth factor activator inhibitor 1 was expressed in Drosophila S2 cells. The recombinant protein was crystallized using ammonium sulfate as precipitant and the crystals diffracted to 3.8 Å resolution, with unit-cell parameters a = b = 95.42, c = 124.50 Å.
To explore the sequence–structure relationships of four-α-helical bundles, the revRM6 protein, which has been engineered with the exact inverse amino-acid sequence of a hyperthermophilic helical bundle, was expressed, purified and crystallized and X-ray diffraction data were collected.
Open  access
access
 access
accessThe crystal structure of SP0092 was determined at 1.61 Å resolution and reveals a domain-swapped dimer with the monomer subunit in a closed conformation in the absence of ligand.
PDB reference: SP0092, 5mlt


 journal menu
journal menu
















![[publBio]](/logos/publbio.gif)